Содержание
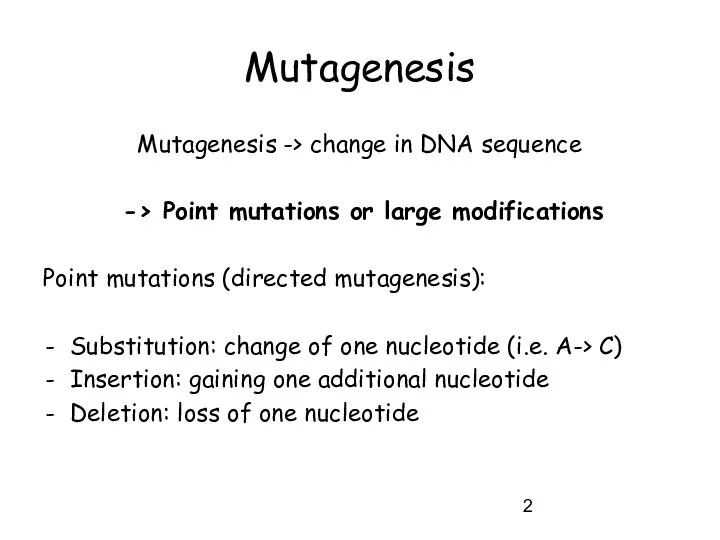
- 2. Mutagenesis Mutagenesis -> change in DNA sequence -> Point mutations or large modifications Point mutations (directed
- 3. Consequences of point mutations within a coding sequence (gene) for the protein Silent mutations: -> change
- 4. Mutagenesis Comparison of cellular and invitro mutagenesis
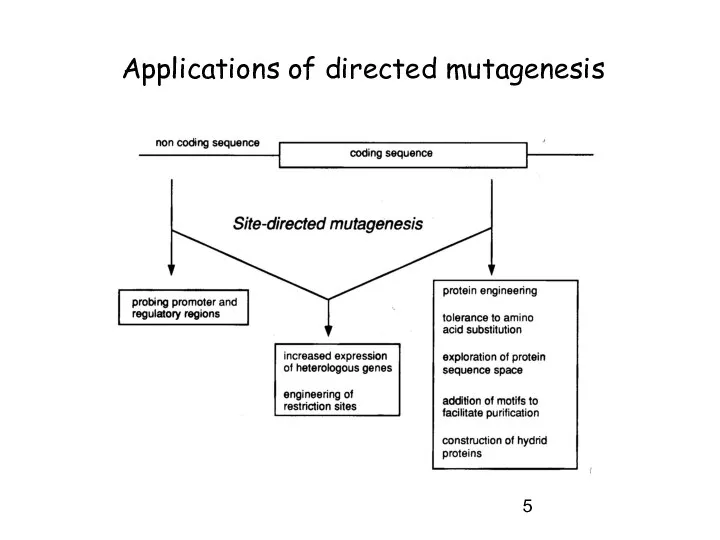
- 5. Applications of directed mutagenesis
- 6. General strategy for directed mutagenesis Requirements: DNA of interest (gene or promoter) must be cloned Expression
- 7. Approaches for directed mutagenesis -> site-directed mutagenesis -> point mutations in particular known area result ->
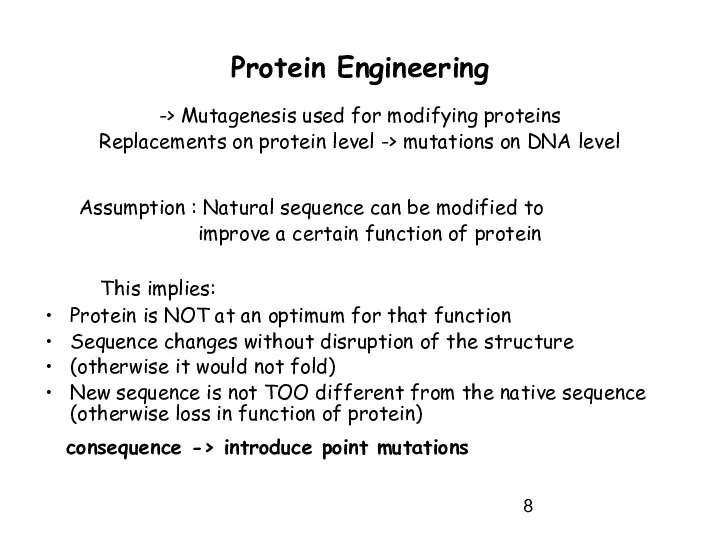
- 8. Protein Engineering -> Mutagenesis used for modifying proteins Replacements on protein level -> mutations on DNA
- 9. Protein Engineering Obtain a protein with improved or new properties
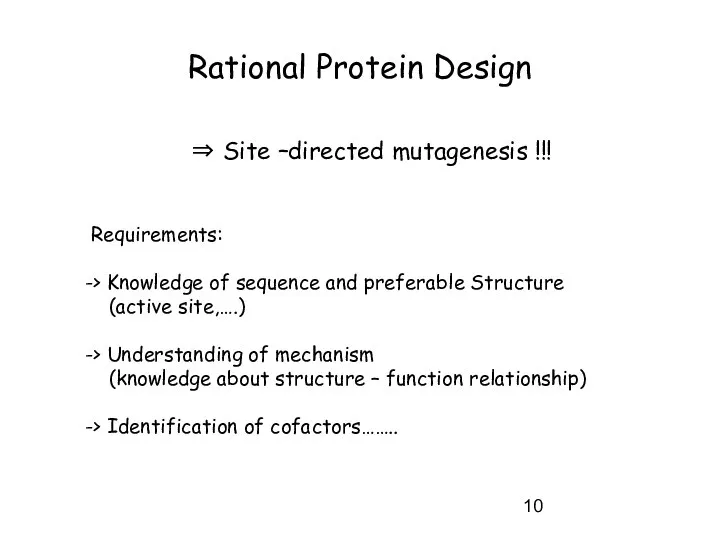
- 10. Rational Protein Design ⇒ Site –directed mutagenesis !!! Requirements: -> Knowledge of sequence and preferable Structure
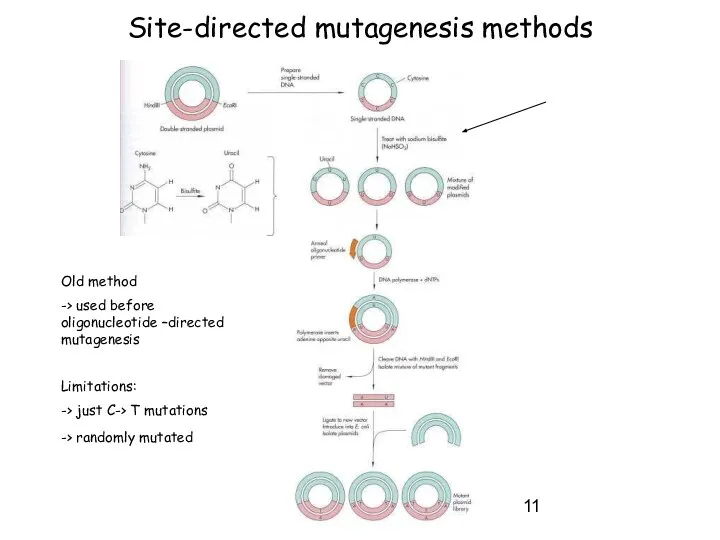
- 11. Site-directed mutagenesis methods Old method -> used before oligonucleotide –directed mutagenesis Limitations: -> just C-> T
- 12. Site-directed mutagenesis methods
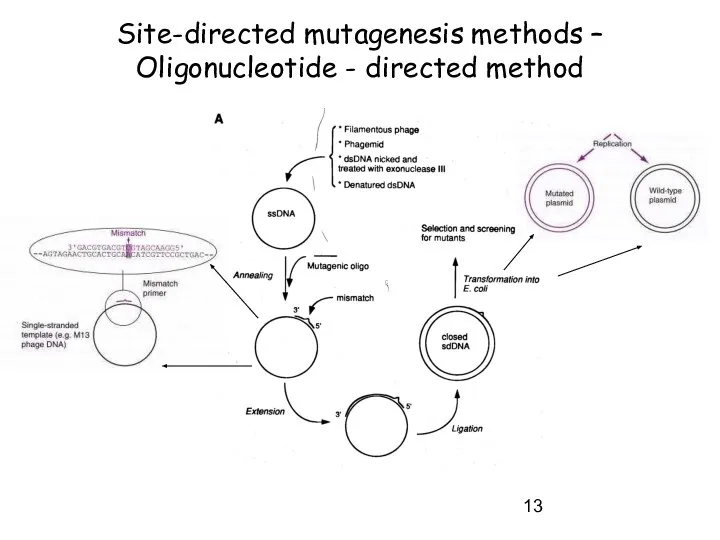
- 13. Site-directed mutagenesis methods – Oligonucleotide - directed method
- 14. Site-directed mutagenesis methods – PCR based
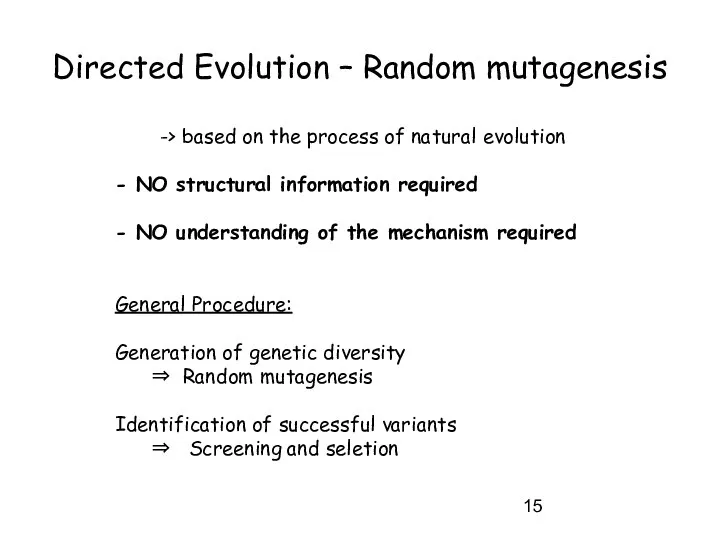
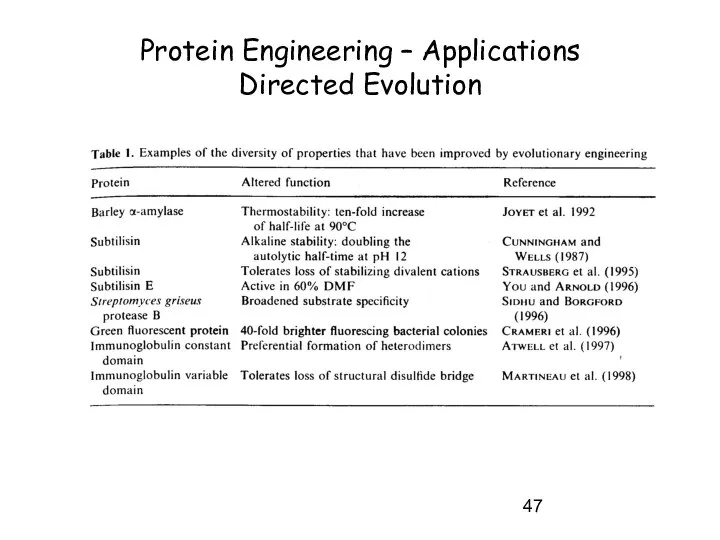
- 15. Directed Evolution – Random mutagenesis -> based on the process of natural evolution - NO structural
- 17. General Directed Evolution Procedure Random mutagenesis methods
- 18. Directed Evolution Library Even a large library -> (108 independent clones) will not exhaustively encode all
- 19. Limitation of Directed Evolution Evolutionary path must exist - > to be successful Screening method must
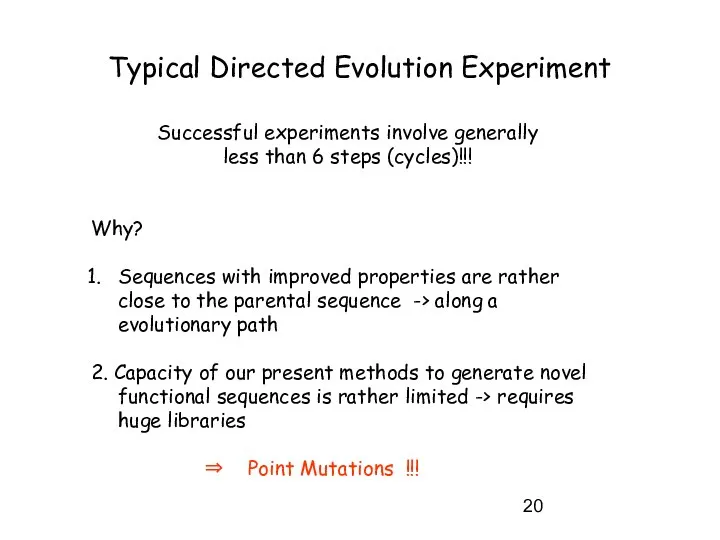
- 20. Successful experiments involve generally less than 6 steps (cycles)!!! Why? Sequences with improved properties are rather
- 21. Evolutionary Methods Non-recombinative methods: -> Oligonucleotide Directed Mutagenesis (saturation mutagenesis) -> Chemical Mutagenesis, Bacterial Mutator Strains
- 22. Evolutionary Methods Type of mutation – Fitness of mutants Type of mutations: Beneficial mutations (good) Neutral
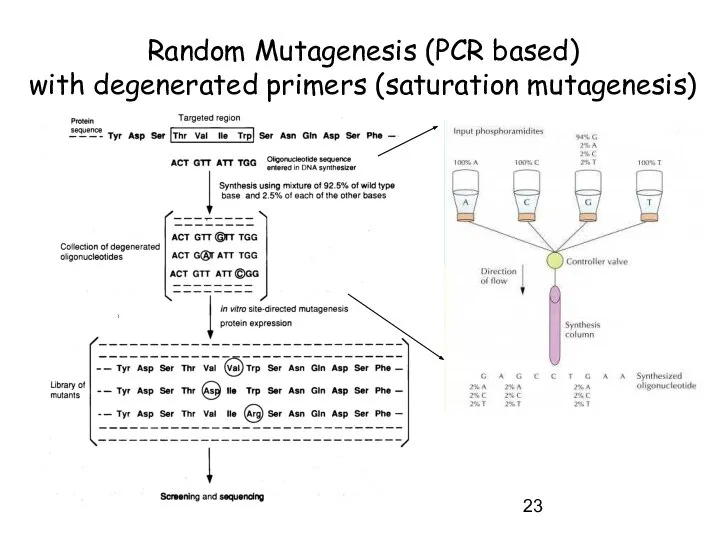
- 23. Random Mutagenesis (PCR based) with degenerated primers (saturation mutagenesis)
- 24. Random Mutagenesis (PCR based) with degenerated primers (saturation mutagenesis)
- 25. Random Mutagenesis (PCR based) Error –prone PCR -> PCR with low fidelity !!! Achieved by: -
- 26. Random Mutagenesis (PCR based) DNA Shuffling DNase I treatment (Fragmentation, 10-50 bp, Mn2+) Reassembly (PCR without
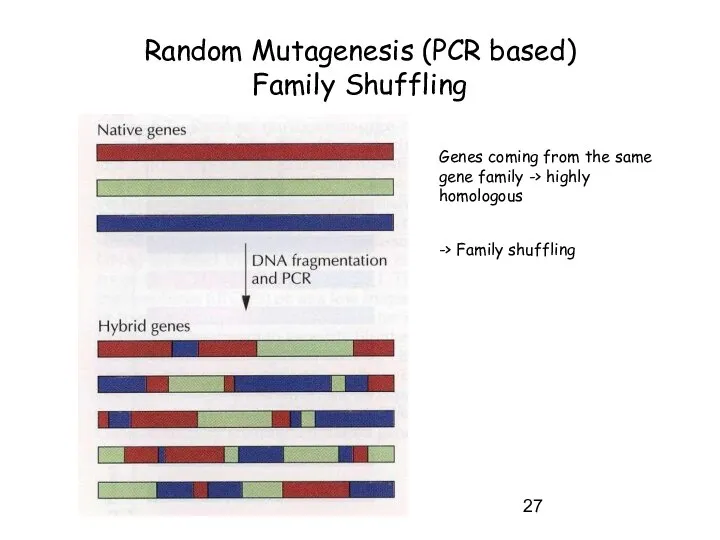
- 27. Random Mutagenesis (PCR based) Family Shuffling Genes coming from the same gene family -> highly homologous
- 28. Random Mutagenesis (PCR based)
- 29. Directed Evolution Difference between non-recombinative and recombinative methods Non-recombinative methods recombinative methods -> hybrids (chimeric proteins)
- 30. Protein Engineering What can be engineered in Proteins ? -> Folding (+Structure): 1. Thermodynamic Stability (Equilibrium
- 31. Protein Engineering What can be engineered in Proteins ? -> Function: 1. Binding (Interaction of a
- 32. Protein Engineering Factors which contribute to stability: Hydrophobicity (hydrophobic core) Electrostatic Interactions: -> Salt Bridges ->
- 33. Protein Engineering Design of Thermal and Environmental stability: Stabilization of α-Helix Macrodipoles Engineer Structural Motifes (like
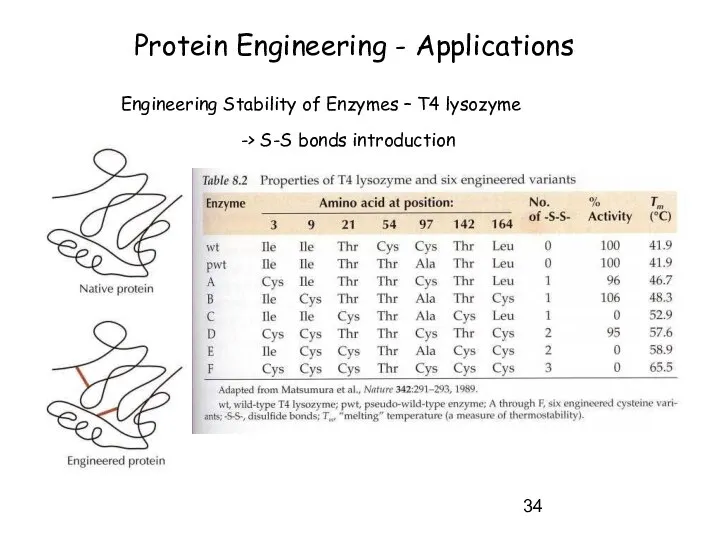
- 34. Protein Engineering - Applications Engineering Stability of Enzymes – T4 lysozyme -> S-S bonds introduction
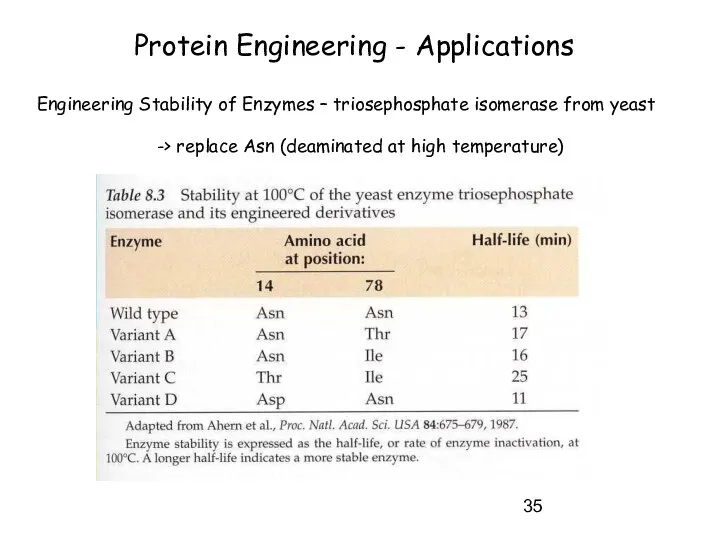
- 35. Protein Engineering - Applications Engineering Stability of Enzymes – triosephosphate isomerase from yeast -> replace Asn
- 36. Protein Engineering - Applications Engineering Activity of Enzymes – tyrosyl-tRNA synthetase from B. stearothermophilus -> replace
- 37. Protein Engineering - Applications Engineering Ca-independency of subtilisin Saturation mutagenesis -> 7 out of 10 regions
- 38. DNA shuffling JCohen. News note: How DNA shuffling works. Sci 293:237 (2001) Maxygen, PCR without synthetic
- 39. Altering multiple properties: rapid high-throughput screening ex., subtilisin Use 26 different subtilisin genes Shuffle DNA, construct
- 40. Laundry, detergent and mushrooms Peroxidase, ink cap mushroom; dye transfer inhibitor Wash conditions: bleach-containing detergents, pH
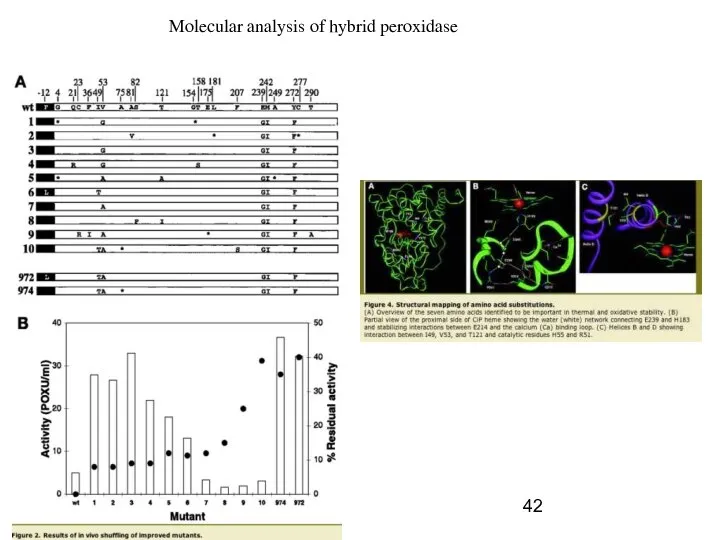
- 41. ex., Coprinus cinereus heme peroxidase (ink cap mushroom); 343 AAc, heme prosthetic group Multiple rounds of
- 42. Molecular analysis of hybrid peroxidase
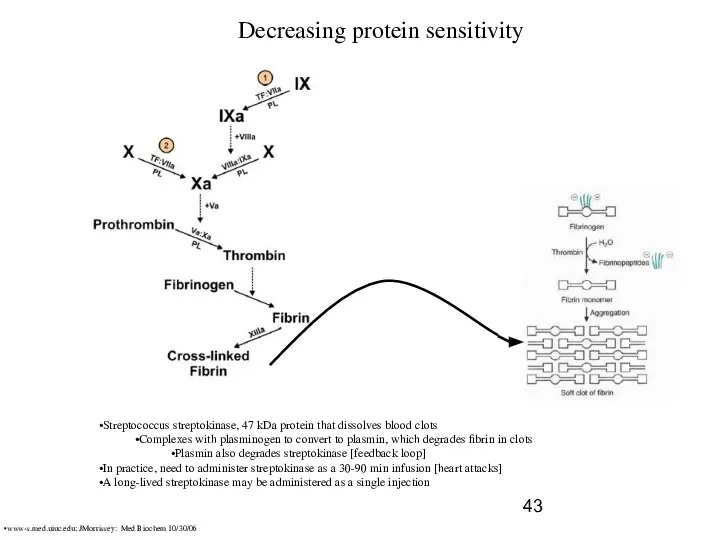
- 43. Decreasing protein sensitivity Streptococcus streptokinase, 47 kDa protein that dissolves blood clots Complexes with plasminogen to
- 44. Decreasing protein sensitivity Streptococcus streptokinase, plasmin sensitivity domain Attacks at Lys59 and Lys382, near each end
- 45. Protein Engineering - Applications Site-directed mutagenesis -> used to alter a single property Problem : changing
- 46. Protein Engineering – Applications Directed Evolution
- 47. Protein Engineering – Applications Directed Evolution
- 48. Protein Engineering – Applications Directed Evolution
- 49. Protein Engineering – Applications Directed Evolution
- 50. Protein Engineering – Directed Evolution
- 51. Protein Engineering - Applications
- 53. Скачать презентацию



































 The life of William Shakespeare
The life of William Shakespeare  Словообразование при помощи префиксов и суффиксов
Словообразование при помощи префиксов и суффиксов Sleepy Animals
Sleepy Animals 10 worst catastrophes in the world!
10 worst catastrophes in the world!  San Francisco The presentation made by Tanya Bezkrovna
San Francisco The presentation made by Tanya Bezkrovna  Sports
Sports Презентация по теме «Изобретатели и изобретения» подготовлена Рогожкиной Кариной Андреевной INVENTIONS AND INVENTORS
Презентация по теме «Изобретатели и изобретения» подготовлена Рогожкиной Кариной Андреевной INVENTIONS AND INVENTORS Types of schools
Types of schools Diet To be or not to be?
Diet To be or not to be?  Be going to
Be going to Презентация по английскому языку Pages of History. Linking Past and Present. Страницы истории. Связь прошлого и настоящего.
Презентация по английскому языку Pages of History. Linking Past and Present. Страницы истории. Связь прошлого и настоящего.  Summer Fun
Summer Fun Изготовление сувенира «Плетень»
Изготовление сувенира «Плетень»  Shopping as an art or art as a shopping
Shopping as an art or art as a shopping Black Square Kazimir Malevich Prepared by Oksana Dubinska Form 11 A School №3
Black Square Kazimir Malevich Prepared by Oksana Dubinska Form 11 A School №3  Игра-викторина Знаете ли вы английский?
Игра-викторина Знаете ли вы английский? Los Angeles Julia Koval Form 11-A
Los Angeles Julia Koval Form 11-A  Презентация Хорошие манеры
Презентация Хорошие манеры My hometown
My hometown Презентация к уроку английского языка "Cover Letter" - скачать бесплатно
Презентация к уроку английского языка "Cover Letter" - скачать бесплатно Defining applied our goals, objectives, and research opportunities
Defining applied our goals, objectives, and research opportunities Parts of the body
Parts of the body Highland games
Highland games  Past Tenses
Past Tenses Презентация к уроку английского языка "Holidays in Canada" - скачать
Презентация к уроку английского языка "Holidays in Canada" - скачать  Your Title Here
Your Title Here M
M Deutschland. Ein Wintermärchen
Deutschland. Ein Wintermärchen