Initiation of translation in prokaryotes: initiation factors, initiator codons, 3'end of RNA small ribosomal subunit
Содержание
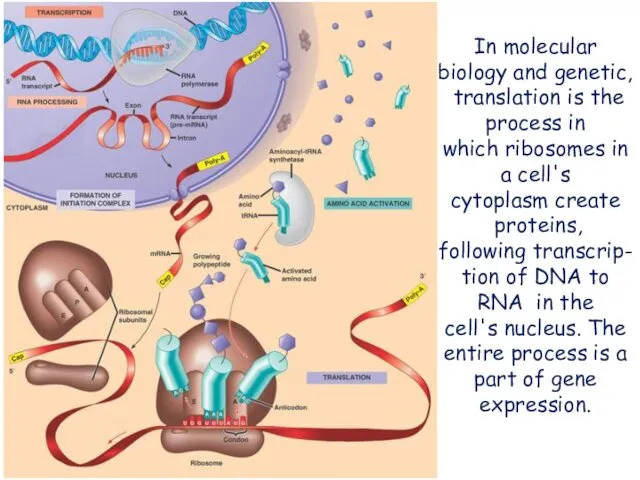
- 2. In molecular biology and genetic, translation is the process in which ribosomes in a cell's cytoplasm
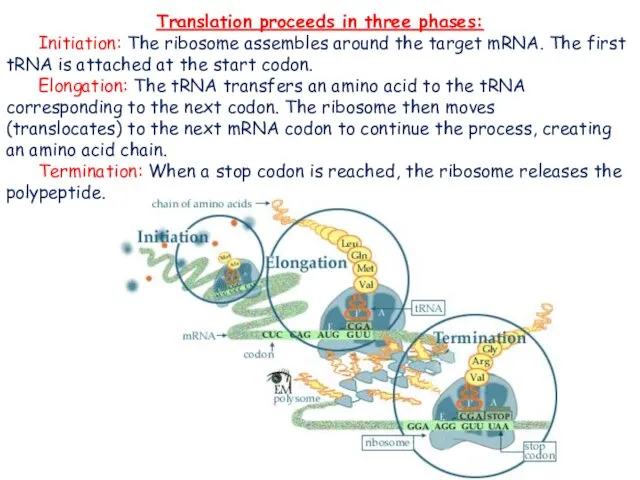
- 3. Translation proceeds in three phases: Initiation: The ribosome assembles around the target mRNA. The first tRNA
- 4. In bacteria, translation occurs in the cytoplasm, where the large and small subunits of the ribosome
- 5. A number of antibiotics act by inhibiting translation. These include anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin,
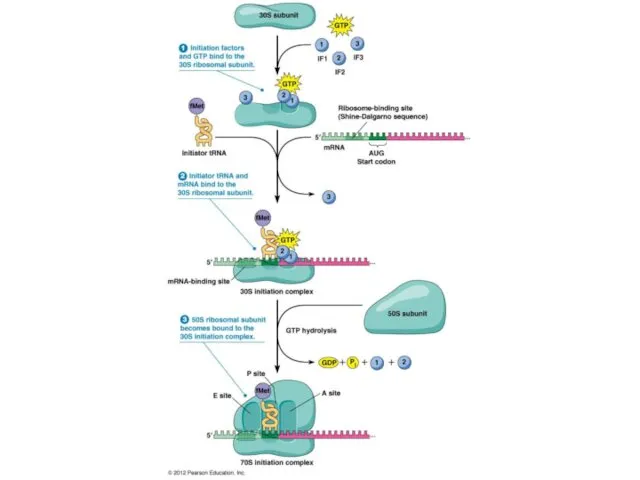
- 6. Translation initiation: Initiation factors Prokaryotes require the use of three initiation factors: IF1, IF2, and IF3,
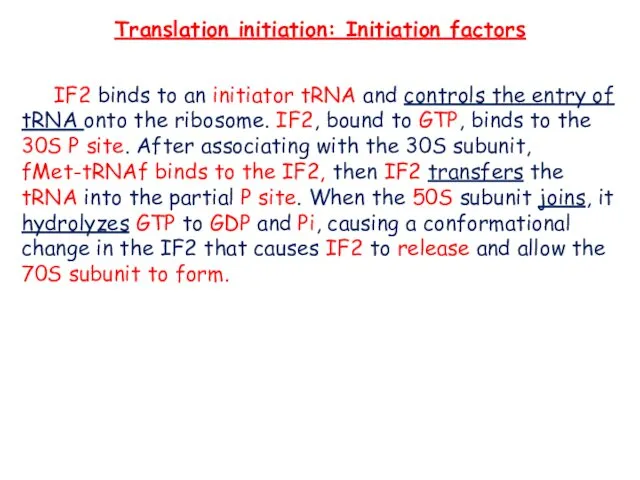
- 7. Translation initiation: Initiation factors IF2 binds to an initiator tRNA and controls the entry of tRNA
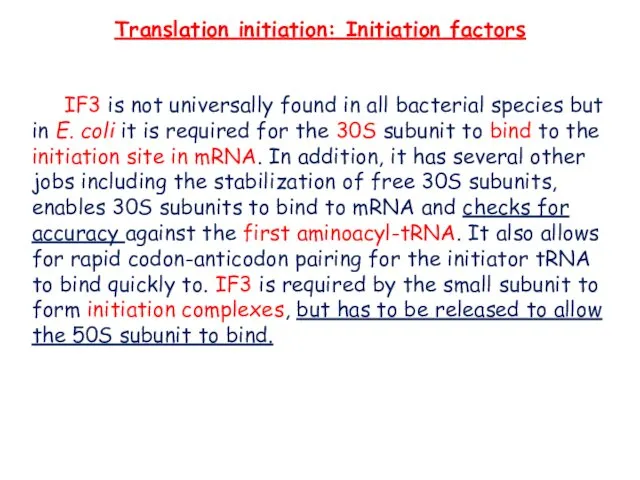
- 8. Translation initiation: Initiation factors IF3 is not universally found in all bacterial species but in E.
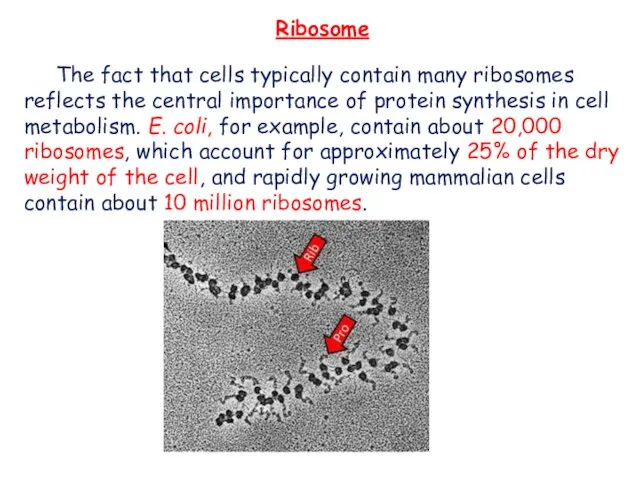
- 10. The fact that cells typically contain many ribosomes reflects the central importance of protein synthesis in
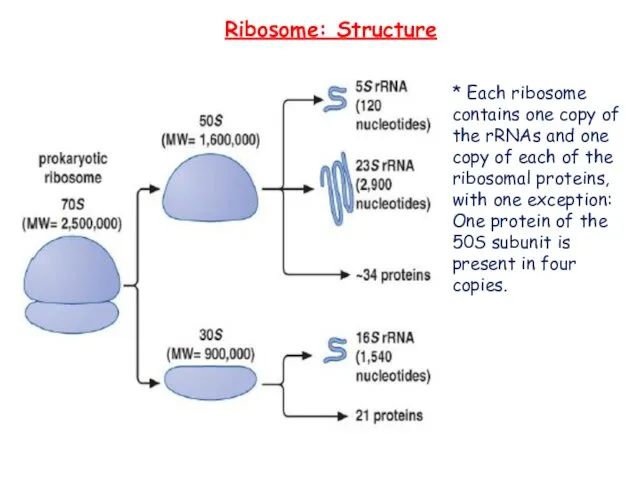
- 11. * Each ribosome contains one copy of the rRNAs and one copy of each of the
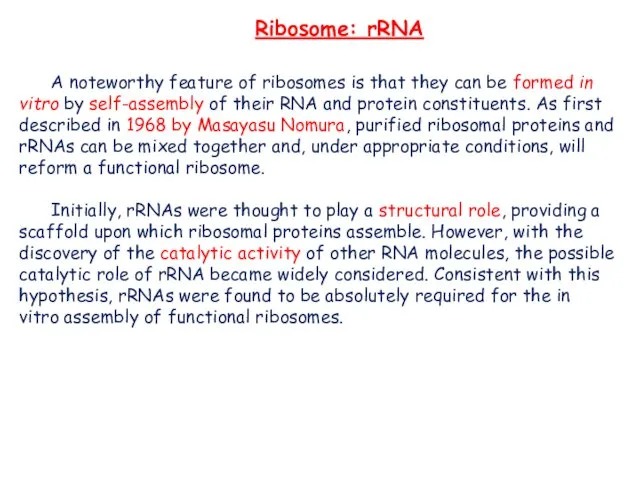
- 12. A noteworthy feature of ribosomes is that they can be formed in vitro by self-assembly of
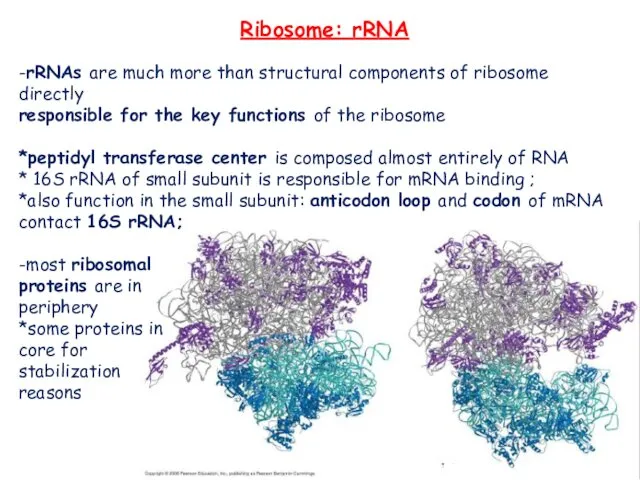
- 13. Ribosome: rRNA -rRNAs are much more than structural components of ribosome directly responsible for the key
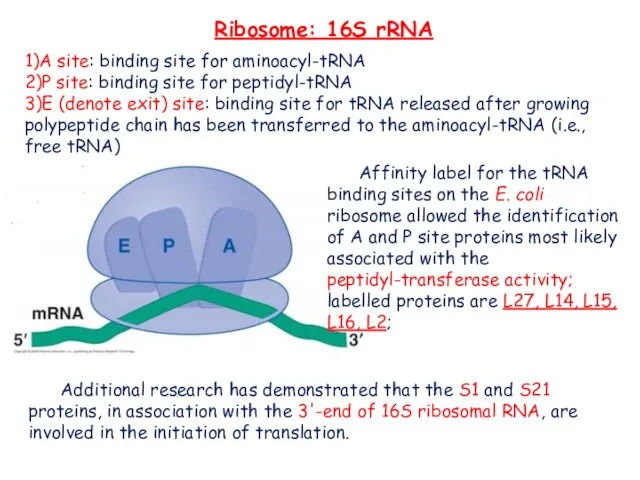
- 14. Ribosome: 16S rRNA 1)A site: binding site for aminoacyl-tRNA 2)P site: binding site for peptidyl-tRNA 3)E
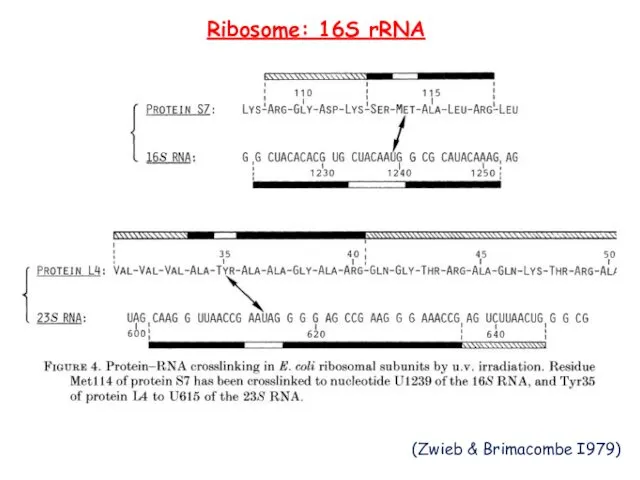
- 15. Ribosome: 16S rRNA (Zwieb & Brimacombe I979)
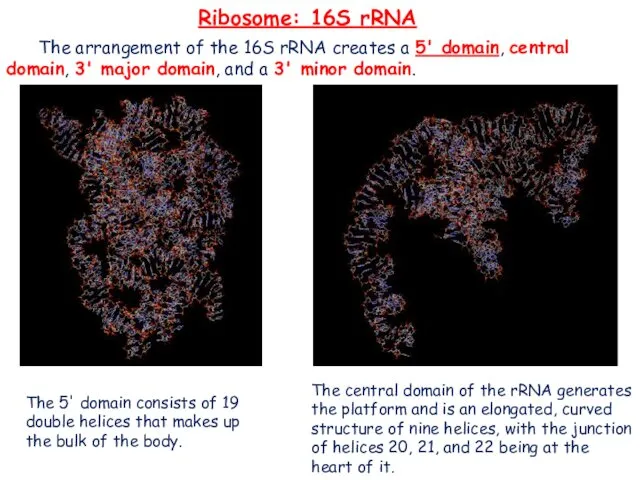
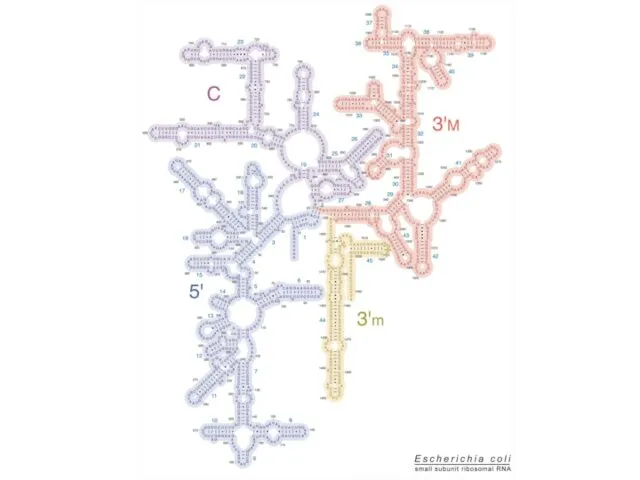
- 16. Ribosome: 16S rRNA The arrangement of the 16S rRNA creates a 5' domain, central domain, 3'
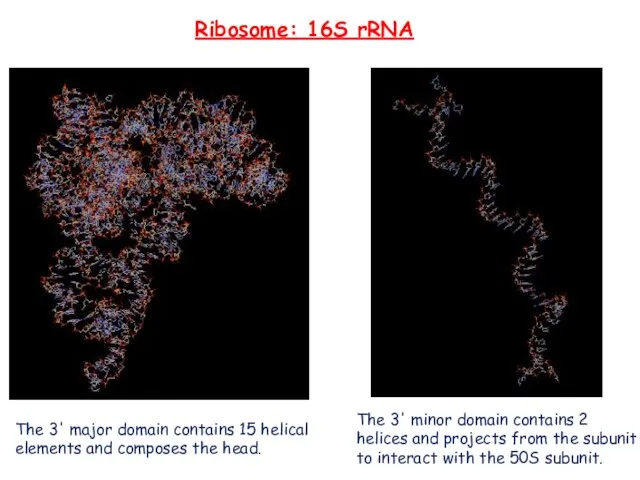
- 17. Ribosome: 16S rRNA The 3' major domain contains 15 helical elements and composes the head. The
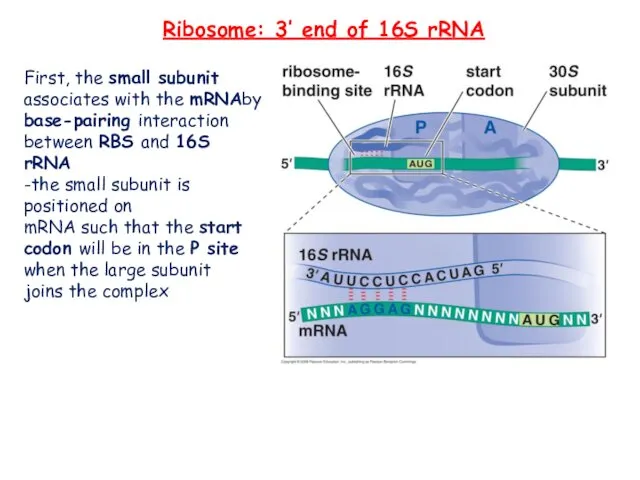
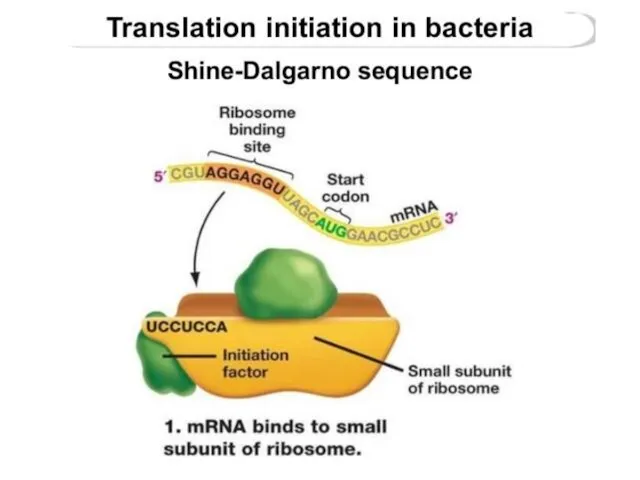
- 19. First, the small subunit associates with the mRNAby base-pairing interaction between RBS and 16S rRNA -the
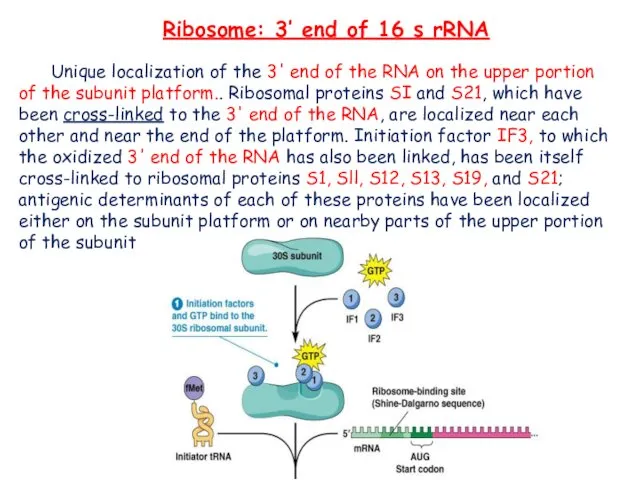
- 20. Ribosome: 3’ end of 16 s rRNA Unique localization of the 3' end of the RNA
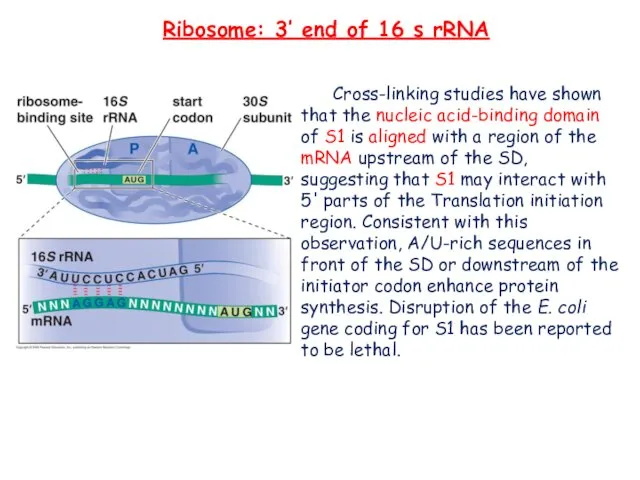
- 21. Ribosome: 3’ end of 16 s rRNA Cross-linking studies have shown that the nucleic acid-binding domain
- 22. Antibiotics affecting 16S rRNA Colicin E3 (protein antibiotic from E.coli) makes a single cut in the
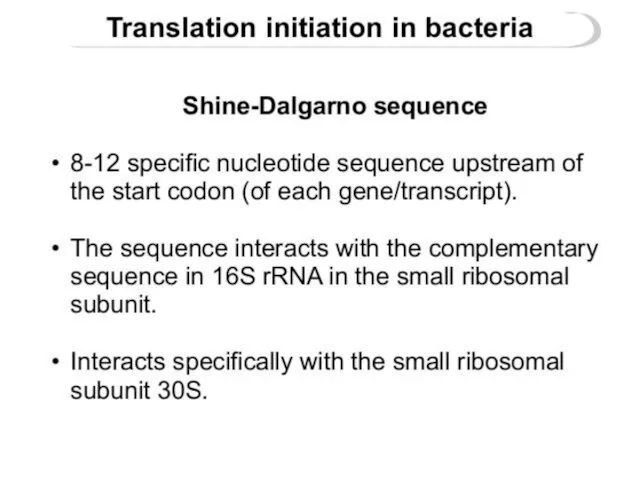
- 23. Shine-Dalgarno sequence
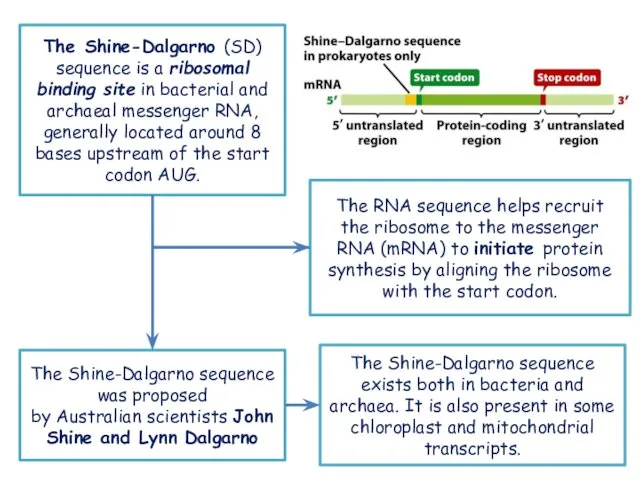
- 24. The Shine-Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally
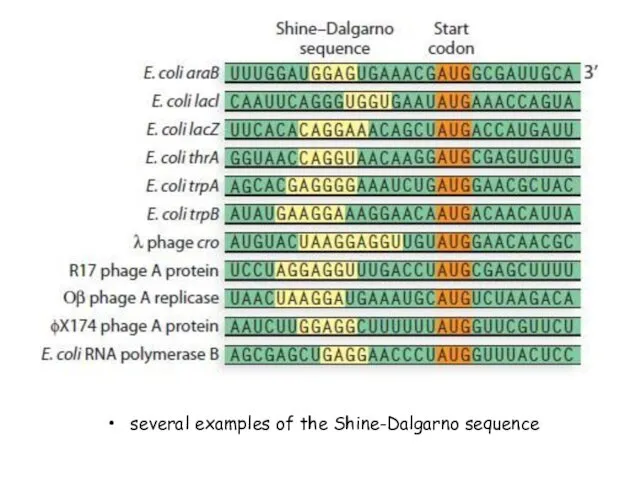
- 27. several examples of the Shine-Dalgarno sequence
- 28. REFERENCE The Cell: A Molecular Approach. 2nd edition.Cooper GM. Sunderland (MA): Sinauer Associates; 2000. Watson J
- 30. Скачать презентацию



 Урок английского языка для 6 класса по учебнику К.И Кауфман, М.Ю.Кауфман Happy English.ru We are fond of pets Учитель: Корнилова Наталья Геннадье
Урок английского языка для 6 класса по учебнику К.И Кауфман, М.Ю.Кауфман Happy English.ru We are fond of pets Учитель: Корнилова Наталья Геннадье Seasons and weather
Seasons and weather PRESENTATION FOR THE STUDENTS OF THE 2nd FORM The teacher : Kudra E.M CLOTHES
PRESENTATION FOR THE STUDENTS OF THE 2nd FORM The teacher : Kudra E.M CLOTHES Opinion essay for the Russian state exam
Opinion essay for the Russian state exam Georgia
Georgia The passive voice
The passive voice Plural number of nouns
Plural number of nouns The Greatest writer in History.
The Greatest writer in History. Презентация к уроку английского языка "Tenses" - скачать бесплатно
Презентация к уроку английского языка "Tenses" - скачать бесплатно Conformity Quotes
Conformity Quotes Russia is my motherland Автор: Гаврилова Елена Валерьевна, учитель английского языка МАОУ СОШ №9 г. Боровичи
Russia is my motherland Автор: Гаврилова Елена Валерьевна, учитель английского языка МАОУ СОШ №9 г. Боровичи  Presentation Anastasia Semko Form 9 School of Podilia
Presentation Anastasia Semko Form 9 School of Podilia  Be not
Be not Deutsche Märchenfilme
Deutsche Märchenfilme  Hardware printer
Hardware printer Презентация к уроку английского языка "Английские заимствования" - скачать бесплатно
Презентация к уроку английского языка "Английские заимствования" - скачать бесплатно Present simple
Present simple Counting
Counting Hippie is a specific subculture
Hippie is a specific subculture Sakura game
Sakura game Do the crossword
Do the crossword My house
My house учащейся 9класса Рыжаковой Анастасии Воспитатель: Яндубаева Екатерина Витальевна 2014г.
учащейся 9класса Рыжаковой Анастасии Воспитатель: Яндубаева Екатерина Витальевна 2014г. The Flag of The United Kingdom
The Flag of The United Kingdom Stroganov Palace Жилюк Екатерина Гр. № 411
Stroganov Palace Жилюк Екатерина Гр. № 411 Discover English
Discover English Презентация к уроку английского языка "All Work and no Play" - скачать
Презентация к уроку английского языка "All Work and no Play" - скачать  Презентация к уроку английского языка "The role of English language in people’s life." - скачать бесплатно
Презентация к уроку английского языка "The role of English language in people’s life." - скачать бесплатно