Содержание
- 2. What is Yeast Genetics? Definition of Genetics in Wikipedia: “Genetics (from Ancient Greek γενετικός genetikos, “genitive”
- 3. This slide was nicked from internet lecture notes of a course held at the Universität München
- 4. Pioneers of yeast genetics Øjvind Winge (1886-1964), Carlsberg laboratory, Kopenhagen: http://www.genetics.org/cgi/content/full/158/1/1 Discovery of alternation of Haplo
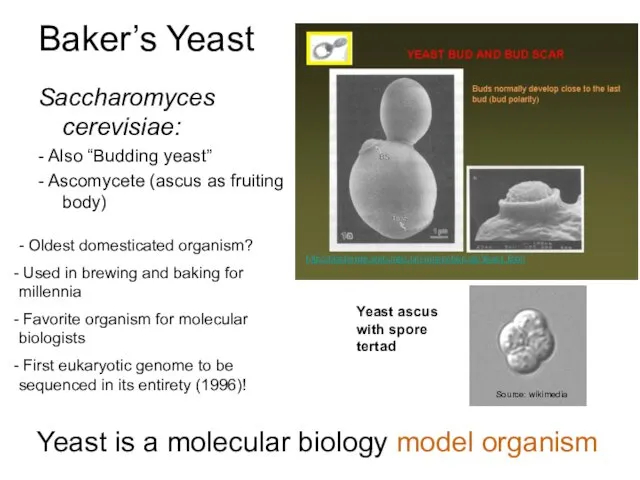
- 5. Baker’s Yeast Saccharomyces cerevisiae: - Also “Budding yeast” - Ascomycete (ascus as fruiting body) - Oldest
- 6. Requirements for Model Organisms:
- 7. Yeast similarity to human cells
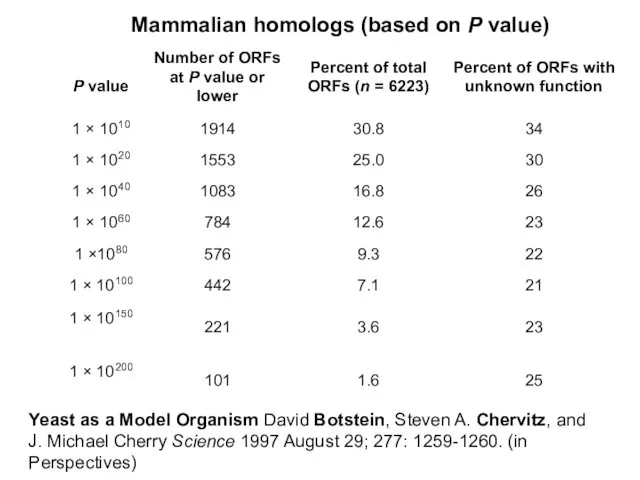
- 8. Yeast as a Model Organism David Botstein, Steven A. Chervitz, and J. Michael Cherry Science 1997
- 9. “Bacterial” aspects of yeast: Single cell organism Haploid growth phase -> phenotype of recessive mutations shows
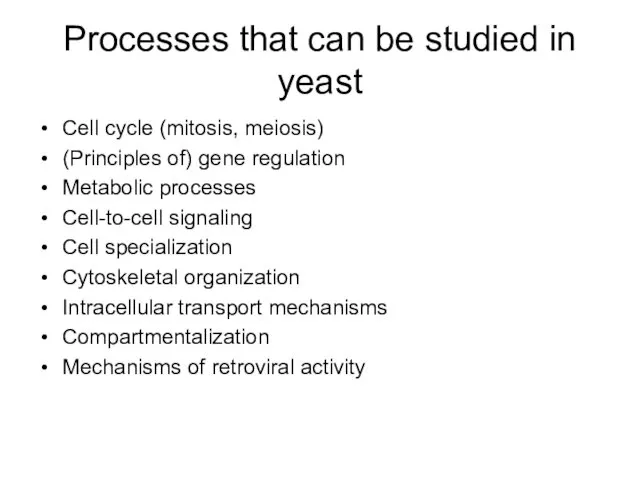
- 10. Processes that can be studied in yeast Cell cycle (mitosis, meiosis) (Principles of) gene regulation Metabolic
- 11. Growth requirements of Baker’s Yeast Wild type S. cerevisiae: prototrophic as long as there is a
- 12. Crabtree effect and oxygen requirements of S. cerevisiae Preferred carbon source: glucose, but many other carbon
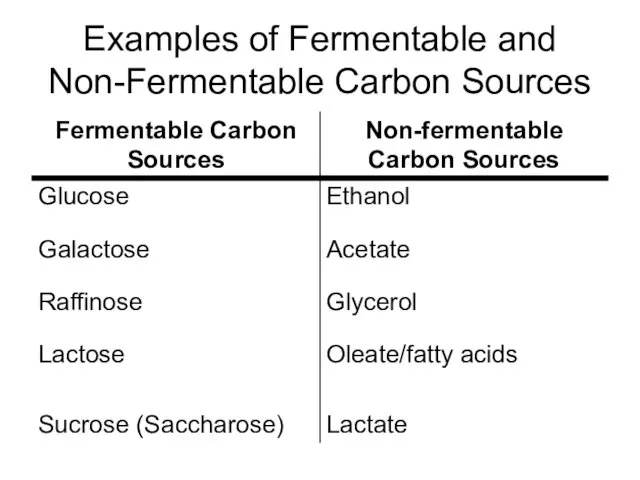
- 13. Examples of Fermentable and Non-Fermentable Carbon Sources
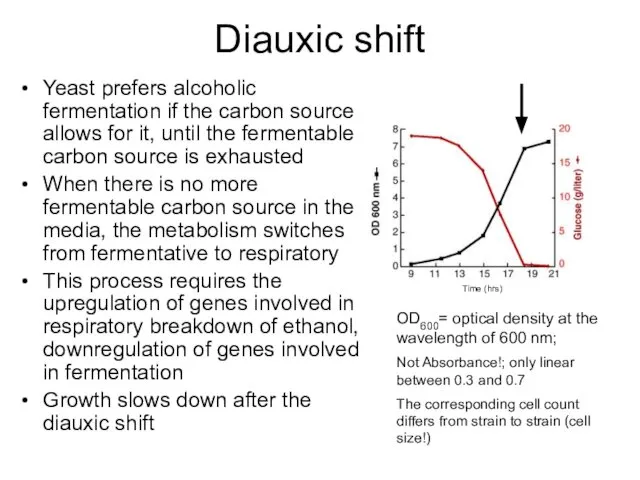
- 14. Diauxic shift Yeast prefers alcoholic fermentation if the carbon source allows for it, until the fermentable
- 15. Growth Media “Favorite” Media (RICH media): YP (Yeast extract and Peptone=peptic digest of meat) + carbon
- 16. Synthetic complete media Contain all the amino acids, some nucleic acid precursors and some vitamins and
- 17. Minimal media Carbon source and Nitrogen source (YNB) Only wild type yeast can grow
- 18. Yeast Gene and Gene Product Nomenclature Dominant alleles are written in italicised capital letters: LEU2, ADE3,
- 20. In most cases the wild type allele is denoted in upper case italics: LEU2, the mutant
- 21. Classical yeast genetics Pre-molecular biology
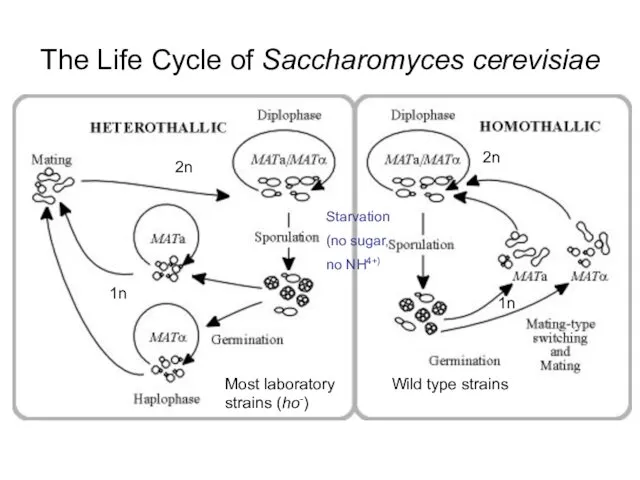
- 23. The Life Cycle of Saccharomyces cerevisiae Wild type strains Most laboratory strains (ho-) 2n 1n 2n
- 24. Yeast has a haploid growth phase Phenotype of mutation apparent immediately Every haploid strain is a
- 25. Genetic Manipulation Ability to mate yeast cells allows combining of mutations Meiotic products (spores) are packed
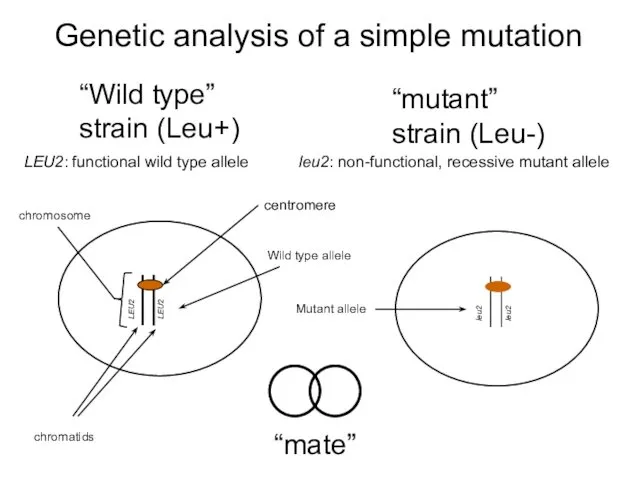
- 26. Genetic analysis of a simple mutation “Wild type” strain (Leu+) “mate”
- 27. Segregation of two alleles involved in Leucine biosynthesis Cells are Leu+, as the functional copy of
- 28. Meiosis 1: separation of the homologous chromosomes Meiosis 2: separation of the chromatids
- 29. Digest off cell wall Tetrad with 3 spores visible in one focal plane and 4th spore
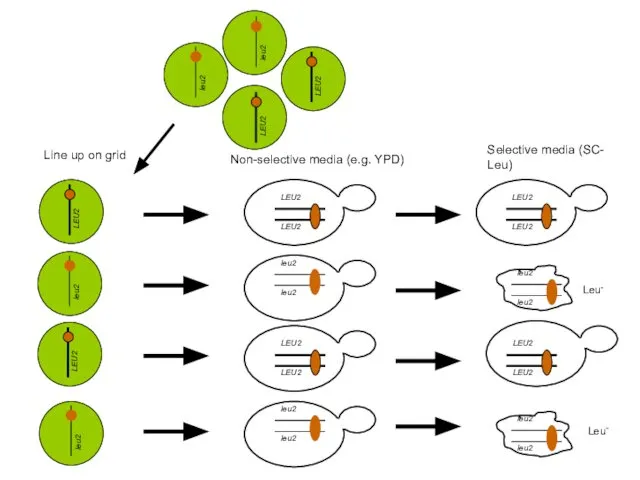
- 31. Line up on grid Non-selective media (e.g. YPD) Selective media (SC- Leu) Leu- Leu-
- 32. Original Dissection on Non-selective plate Replica on selective plate (e.g. Leu- strain on SC – Leucine)
- 33. Segregation of two unlinked genes Example: TRP1, LEU2 Haploid, Leu+, Trp- Haploid, Leu-, Trp+ Diploid, Trp+,
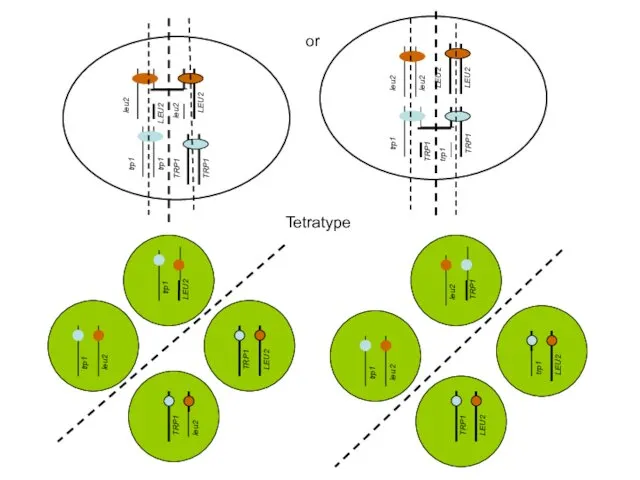
- 34. Possible distribution of chromosomes during meiosis Resulting tetrads after sporulation parental ditype (Trp+, Leu- : Leu+,Trp-)
- 35. or trp1 trp1 TRP1 TRP1 Tetratype
- 36. Ratios of different types of tetrads! (NOT spores)
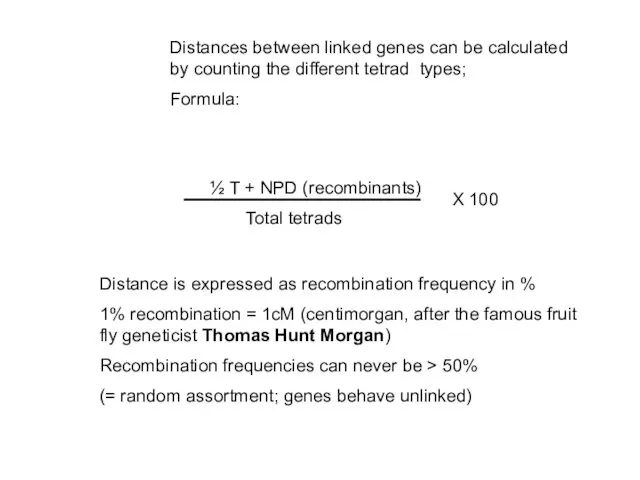
- 37. Distances between linked genes can be calculated by counting the different tetrad types; Formula: ½ T
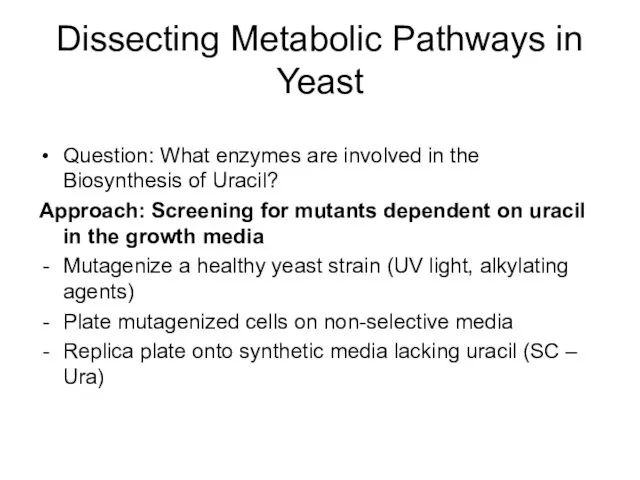
- 38. Dissecting Metabolic Pathways in Yeast Question: What enzymes are involved in the Biosynthesis of Uracil? Approach:
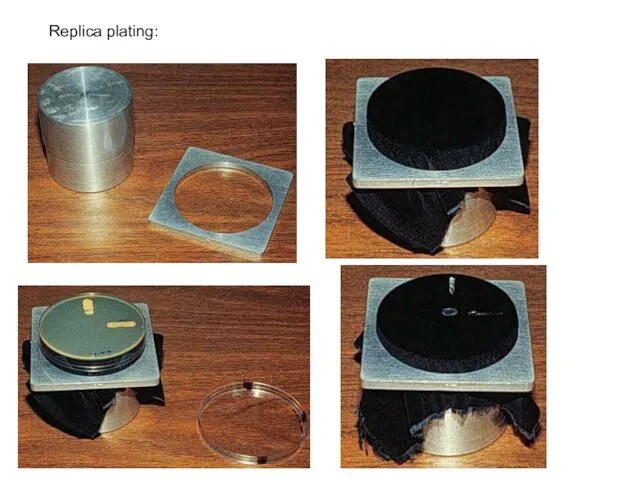
- 39. Replica plating:
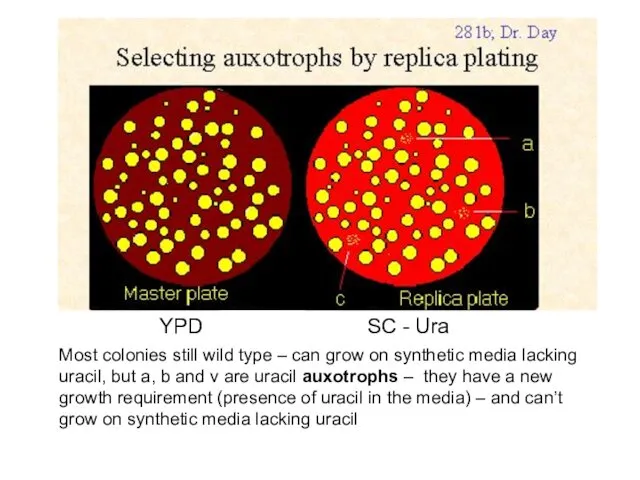
- 40. YPD SC - Ura Most colonies still wild type – can grow on synthetic media lacking
- 41. Sorting of mutations In our hypothetical screen, we have identified several haploid mutants in the uracil
- 42. Complementation analysis Scenario 1: mutations are in the same gene Ura- Ura- Ura- Diploid cannot grow
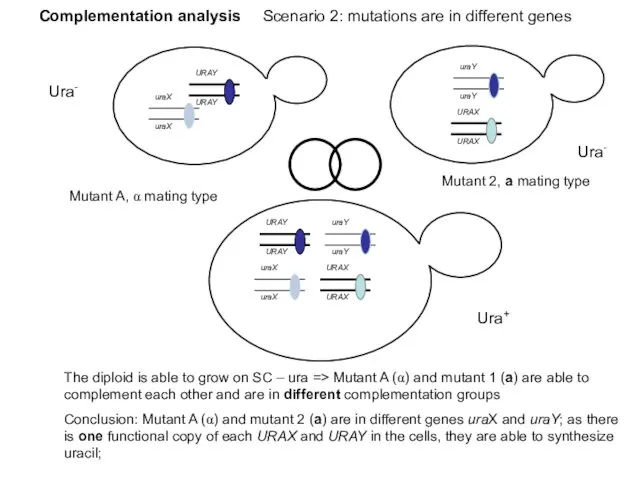
- 43. Complementation analysis Scenario 2: mutations are in different genes Ura- Ura- Ura+ Mutant A, α mating
- 44. Complementation of mutants in the uracil biosynthesis pathway (+) = mutants complement each other ; (-)
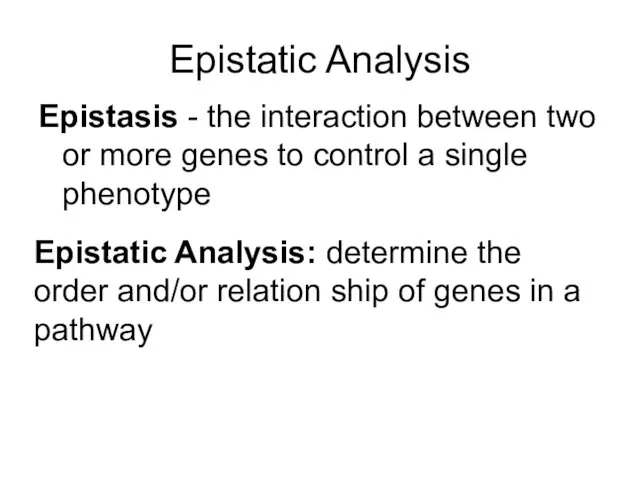
- 45. Epistatic Analysis Epistasis - the interaction between two or more genes to control a single phenotype
- 46. Example of Epistatic analysis Example: Adenine biosynthesis mutants ade2 and ade3 (unlinked genes): ade2 mutants are
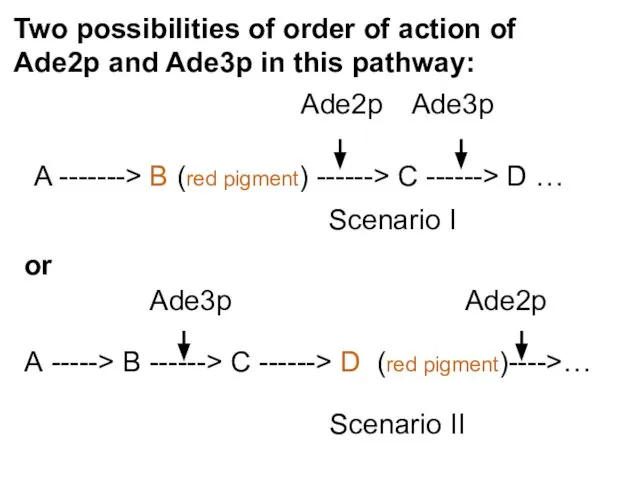
- 47. Scenario I Scenario II Two possibilities of order of action of Ade2p and Ade3p in this
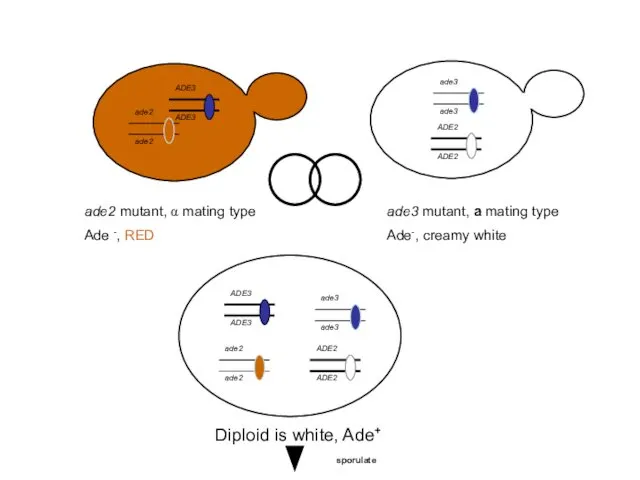
- 48. ade2 mutant, α mating type Ade -, RED ade3 mutant, a mating type Ade-, creamy white
- 49. Possible distribution of chromosomes during meiosis Parental Ditype - uninformative All Ade-
- 50. Nonparental Ditype – informative (two spores carry both mutations) Two Scenarios
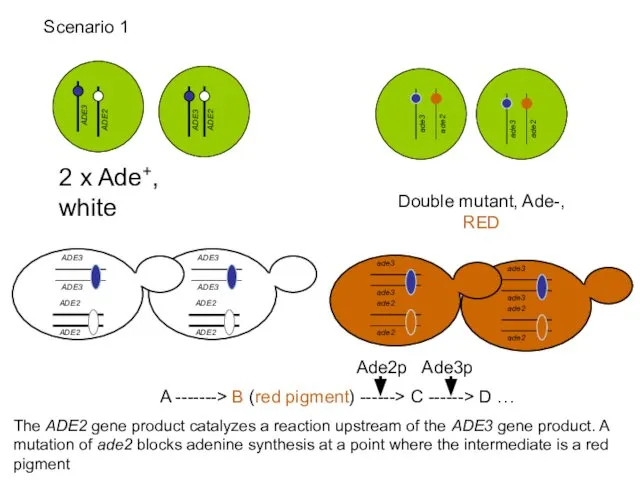
- 51. 2 x Ade+, white Double mutant, Ade-, RED A -------> B (red pigment) ------> C ------>
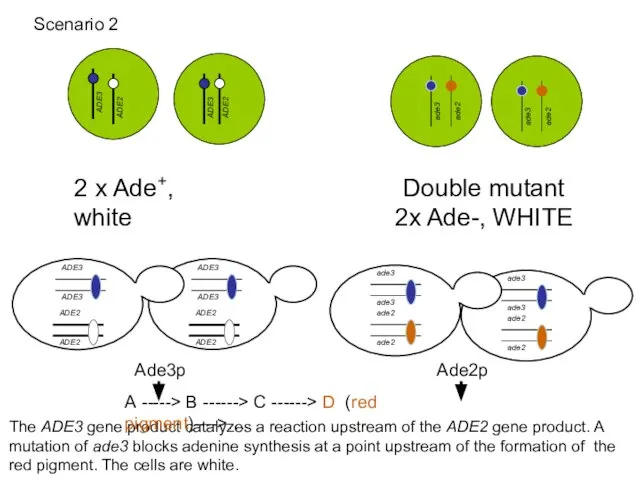
- 52. 2 x Ade+, white Double mutant 2x Ade-, WHITE The ADE3 gene product catalyzes a reaction
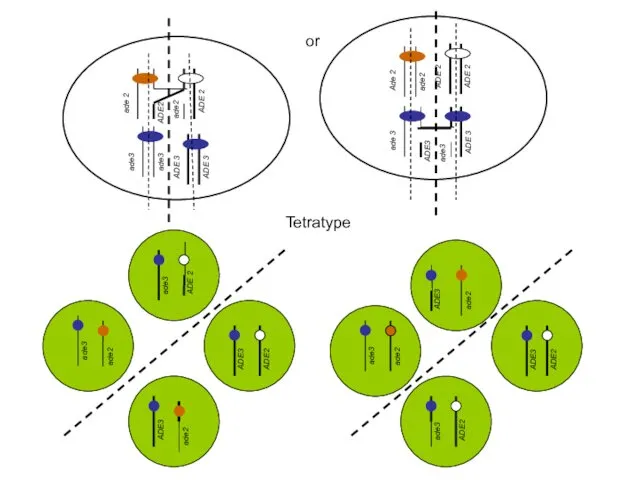
- 53. or ade 3 ade3 ADE3 ADE 3 ADE2 ade 2 ade2 ADE 2 Tetratype
- 54. Tetratype Ade-, white Ade+, white Ade-, red Ade-, white Ade-, white Scenario1 Scenario2 A -----> B
- 56. Скачать презентацию






























 Основи циології
Основи циології КІР Роботу виконала Учениця 10-А класу Ставищенського НВК №2 Христевич Ілона
КІР Роботу виконала Учениця 10-А класу Ставищенського НВК №2 Христевич Ілона  Значение физических упражнений для формирования скелета и мышц
Значение физических упражнений для формирования скелета и мышц Печень, строение и функции
Печень, строение и функции Тварини-будівельники
Тварини-будівельники Cемейства мышиных
Cемейства мышиных Пищеварение. Закономерности пищеварительных процессов
Пищеварение. Закономерности пищеварительных процессов Этапы урока. Организация начала занятия. Проверка выполнения домашнего задания. Подготовка к усвоению новых знаний Изучение
Этапы урока. Организация начала занятия. Проверка выполнения домашнего задания. Подготовка к усвоению новых знаний Изучение  История описательной ботаники XVI-XVIII веков
История описательной ботаники XVI-XVIII веков Агроэкология – основа биологического производства органического вещества
Агроэкология – основа биологического производства органического вещества Степной орёл
Степной орёл Алоэ Төкіш Ақнұр Ерболатқызы №36 орта мектебі
Алоэ Төкіш Ақнұр Ерболатқызы №36 орта мектебі Дарвинизм
Дарвинизм Зимняя жизнь птиц и зверей
Зимняя жизнь птиц и зверей Взаимодействие нервной и эндокринной систем в регуляции генетических процессов
Взаимодействие нервной и эндокринной систем в регуляции генетических процессов Транспортные системы организма
Транспортные системы организма Отряд Стрекозы
Отряд Стрекозы Сокращение скелетных мышц
Сокращение скелетных мышц Садовые цветы - загадки
Садовые цветы - загадки Наводнения. Виды наводнений и их причины. Тема 4.1
Наводнения. Виды наводнений и их причины. Тема 4.1 Закон Харди-Вайнберга
Закон Харди-Вайнберга Оценка влияние гумата Плодородия на урожайность овса в условиях северного Казахстана
Оценка влияние гумата Плодородия на урожайность овса в условиях северного Казахстана Предки человека
Предки человека  Последовательность изучения структуры цветка
Последовательность изучения структуры цветка Генетические алгоритмы
Генетические алгоритмы Гены в хромосомах и популяциях
Гены в хромосомах и популяциях Живой организм. Признаки живых организмов
Живой организм. Признаки живых организмов Закономерности наследования признаков, выявленные Г. Менделем. Законы и опыты Г.Менделя
Закономерности наследования признаков, выявленные Г. Менделем. Законы и опыты Г.Менделя