Содержание
- 4. Nick translation describes the ability of E. coli DNA polymerase I to use a nick as
- 5. The common organization of DNA polymerases has a palm that contains the catalytic site, fingers that
- 6. The crystal structure of phage T7 DNA polymerase shows that the template strand takes a sharp
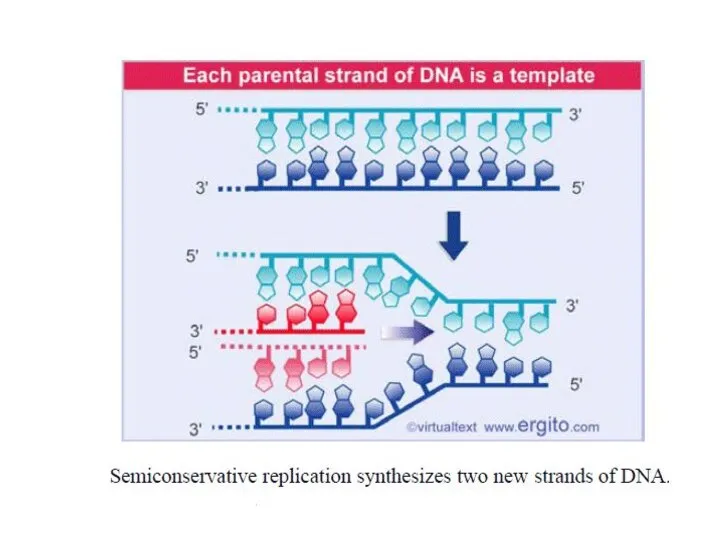
- 7. The leading strand of DNA is synthesized continuously in the 5’-3’ direction. The lagging strand of
- 8. A helicase is an enzyme that uses energy provided by ATP hydrolysis to separate the strands
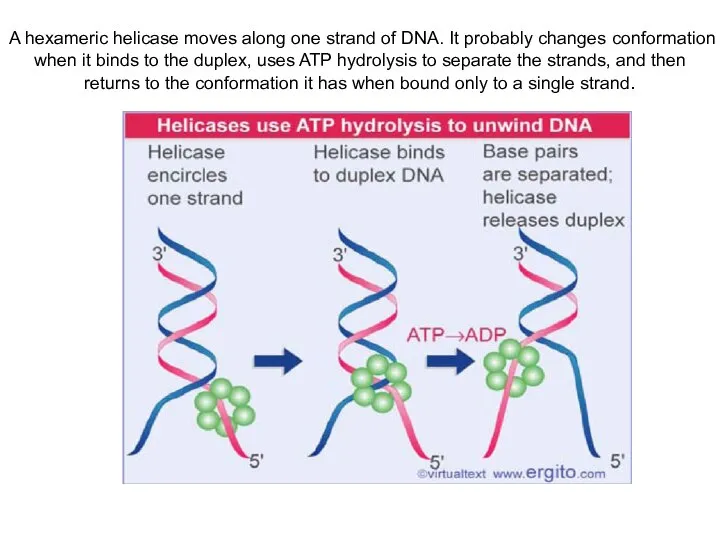
- 9. A hexameric helicase moves along one strand of DNA. It probably changes conformation when it binds
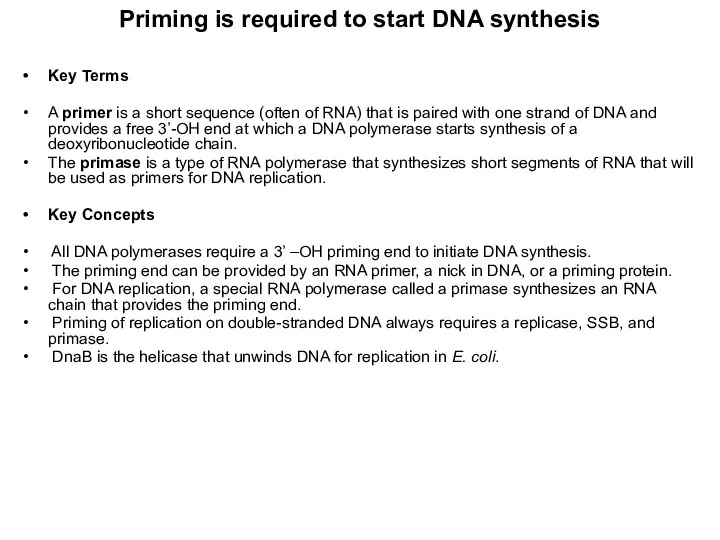
- 10. Priming is required to start DNA synthesis Key Terms A primer is a short sequence (often
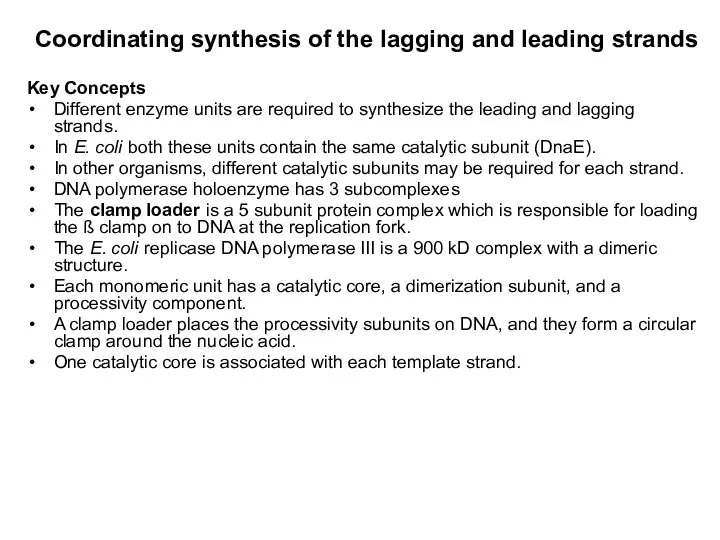
- 11. Coordinating synthesis of the lagging and leading strands Key Concepts Different enzyme units are required to
- 12. DNA polymerase III holoenzyme assembles in stages, generating an enzyme complex that synthesizes the DNA of
- 13. The clamp controls association of core enzyme with DNA · The core on the leading strand
- 17. Okazaki fragments are linked by ligase DNA ligase makes a bond between an adjacent 3’-OH and
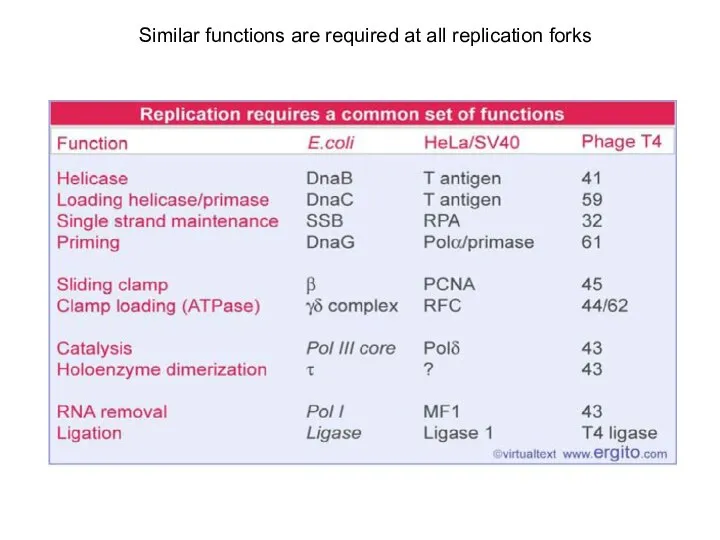
- 18. Similar functions are required at all replication forks
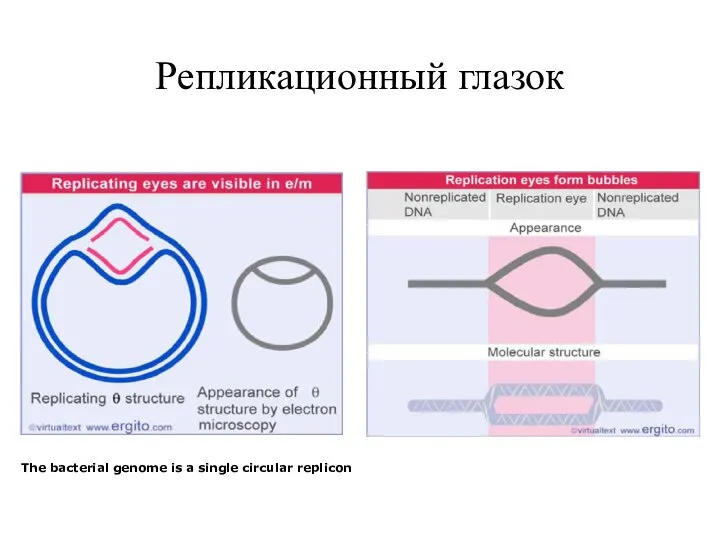
- 19. Репликационный глазок The bacterial genome is a single circular replicon
- 20. Creating the replication forks at an origin The origin of E. coli,oriC, is 245 bp in
- 21. Терминация – встреча вилок
- 23. Скачать презентацию














 Ями на дорогах
Ями на дорогах Циклические алгоритмы. Оператор цикла For.Тело цикла
Циклические алгоритмы. Оператор цикла For.Тело цикла Sonja Gerhardt
Sonja Gerhardt Крест и крестное знамение
Крест и крестное знамение Повторение прошедших тем. Логические выражения
Повторение прошедших тем. Логические выражения Функции элементов нервной системы
Функции элементов нервной системы  Инновационная деятельность в образовании
Инновационная деятельность в образовании  Структура учебно-тренировочного процесса
Структура учебно-тренировочного процесса ГТО – это спорт! Спорт – это жизнь!
ГТО – это спорт! Спорт – это жизнь! коллекция русских икон
коллекция русских икон ТАКТИЧЕСКИЕ ХАРАКТЕРИСТИКИ, ВОЗМОЖНОСТИ И ОСОБЕННОСТИ ЭКСПЛУАТАЦИИ БРОНЕТЕХНИКИ, МОТОЦИКЛОВ, САМОЛЁТОВ И ВЕРТОЛЁТОВ, ИСПОЛЬЗУЕМ
ТАКТИЧЕСКИЕ ХАРАКТЕРИСТИКИ, ВОЗМОЖНОСТИ И ОСОБЕННОСТИ ЭКСПЛУАТАЦИИ БРОНЕТЕХНИКИ, МОТОЦИКЛОВ, САМОЛЁТОВ И ВЕРТОЛЁТОВ, ИСПОЛЬЗУЕМ Синдром жировой эмболии Проф. Н.Е.Буров
Синдром жировой эмболии Проф. Н.Е.Буров Анализ педагогических результатов на основе мониторинга учащихся средней школы ТИНУС ВАЛЕНТИНА МИХАЙЛОВНА учитель физической
Анализ педагогических результатов на основе мониторинга учащихся средней школы ТИНУС ВАЛЕНТИНА МИХАЙЛОВНА учитель физической Азбука архитектуры
Азбука архитектуры Презентация Фотографические принадлежности. Характеристика, классификация, ассортимент
Презентация Фотографические принадлежности. Характеристика, классификация, ассортимент Аналоговые элементы
Аналоговые элементы Noel et Nouvel An en France. Traditions et coutumes
Noel et Nouvel An en France. Traditions et coutumes Единый урок прав человека
Единый урок прав человека Морфофункциональные особенности строения брюшины
Морфофункциональные особенности строения брюшины Земельные, водные и биологические ресурсы России Подготовила Студентка группы Э122б Арсёнова В.
Земельные, водные и биологические ресурсы России Подготовила Студентка группы Э122б Арсёнова В. Реорганизация структурных подразделений ОТ, ТБ и ООС КПО
Реорганизация структурных подразделений ОТ, ТБ и ООС КПО Формы правления. Формы государственно-территориального устройства. Политические режимы
Формы правления. Формы государственно-территориального устройства. Политические режимы Народные промыслы Центрального района России
Народные промыслы Центрального района России К доске
К доске Предмет исторической
Предмет исторической  ангиология неврология
ангиология неврология 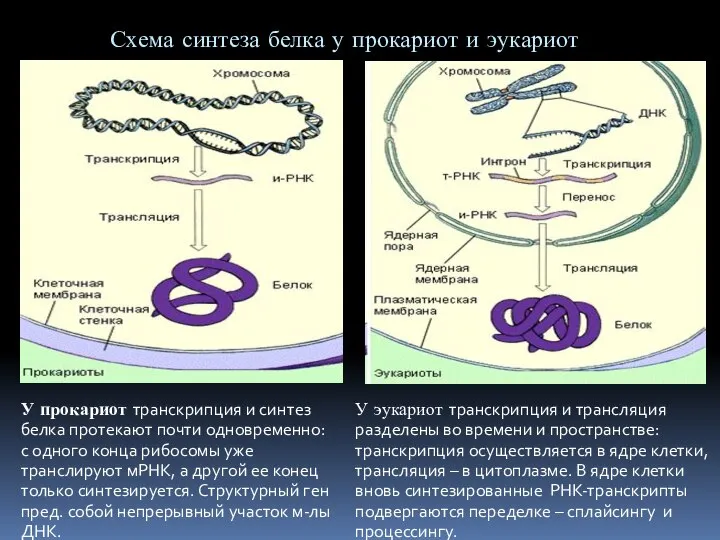 Регуляция синтеза белка
Регуляция синтеза белка Структура команд и режимы адресации на примере PDP-11
Структура команд и режимы адресации на примере PDP-11