Quantitative flow cytometry. Advancing the ability of flow cytometry to serve clinical and research purposes
Содержание
- 2. My story : Biysk
- 3. My story : Novosibirsk Biysk
- 4. My story : Novosibirsk B.S. and M.S. in Physics
- 5. My story : Novosibirsk Ph.D. in Physics and Mathematics
- 6. My story : Novosibirsk Ph.D. in Physics and Mathematics Brno Ph.D. in Biophysics
- 7. My story : Novosibirsk Brno Stanford, CA
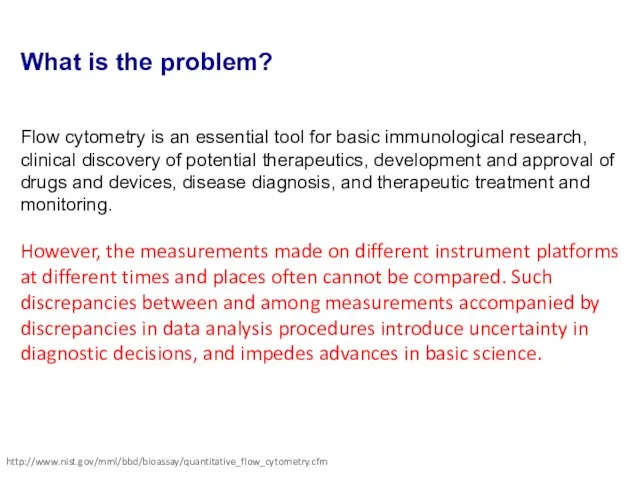
- 8. What is the problem? Flow cytometry is an essential tool for basic immunological research, clinical discovery
- 9. To develop methods and procedures to enable quantitative measurements of biological substances such as cells, proteins,
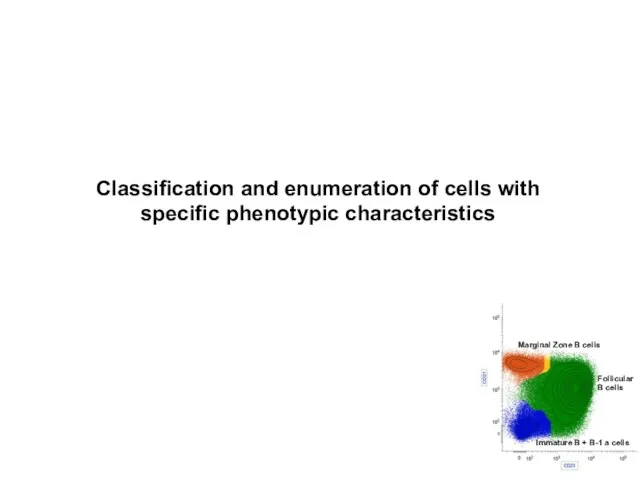
- 10. Accurate classification and enumeration of cells with specific phenotypic characteristics. Quantitation of expression levels of surface
- 11. Classification and enumeration of cells with specific phenotypic characteristics Marginal Zone B cells Follicular B cells
- 12. Schematic of the analysis package that statistical procedures are embedded http://cytogenie.org/
- 13. Projection pursuit seeks one projection at a time http://www.few.vu.nl/~tvpham/images/ppde.jpg http://slideplayer.com/slide/4970323/# Projection Pursuit
- 14. why: Curse of dimensionality Less Robustness Required number of events increases with dimensionality Greater computational cost
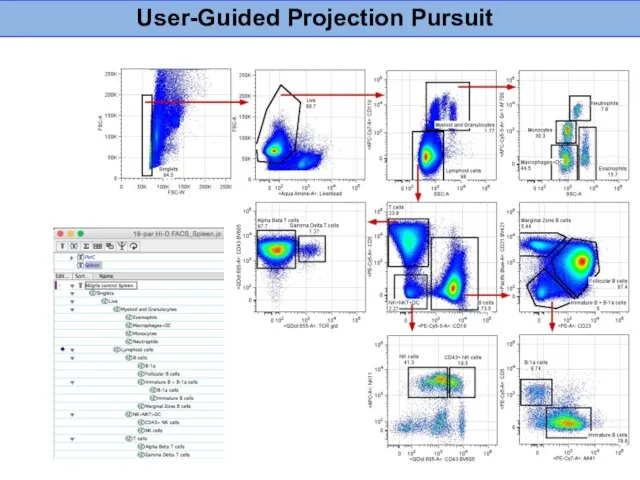
- 15. User-Guided Projection Pursuit
- 16. Walther G. et al, Adv Bioinformatics, 2009 Finding clusters by density based merging (DBM)
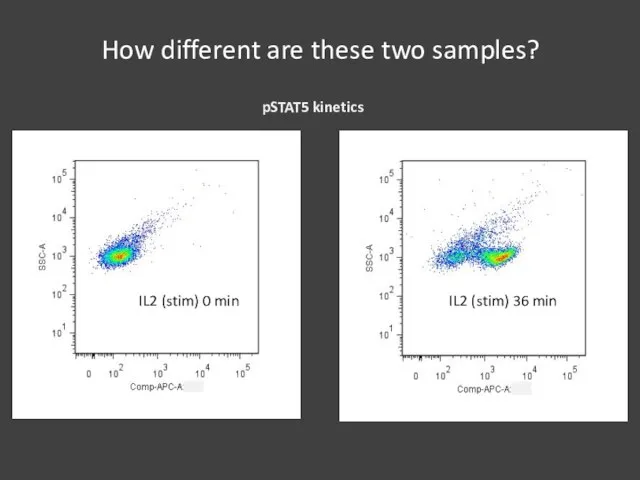
- 18. How different are these two samples?
- 19. The index should: possess the properties of a metric: non-negativity d(x,y) ≥ 0 identity of indiscernibles
- 20. Some test statistics are limited to univariate data, e.g., Kolmogorov-Smirnoff statistic and Overton Subtraction (Sheskin, 2000)
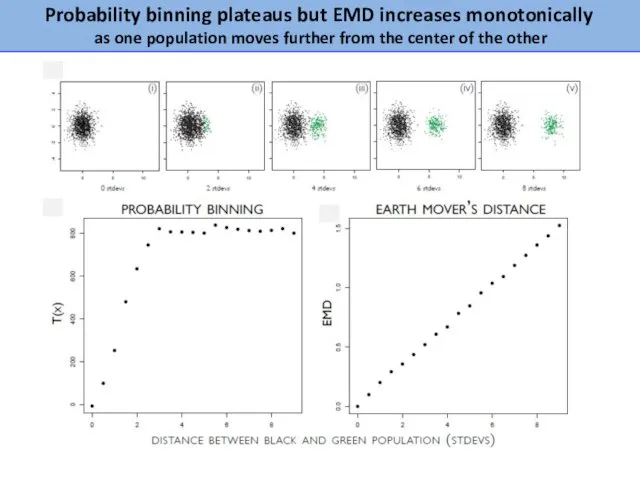
- 21. Probability binning plateaus but EMD increases monotonically as one population moves further from the center of
- 22. EMD is the minimum cost of turning one pile of dirt into the other where the
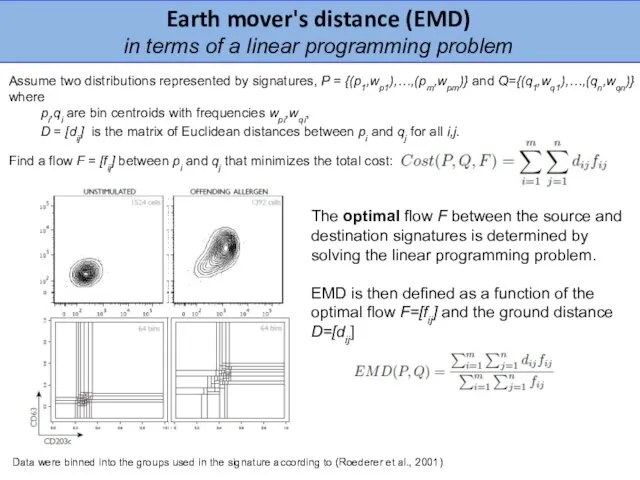
- 23. Assume two distributions represented by signatures, P = {(p1,wp1),…,(pm,wpm)} and Q={(q1,wq1),…,(qn,wqn)} where pi,qi are bin centroids
- 24. Diagnostic tool for distinguishing cystic fibrosis (CF) from allergic bronchopulmonary aspergillosis (ABPA) in CF Surface CD203c
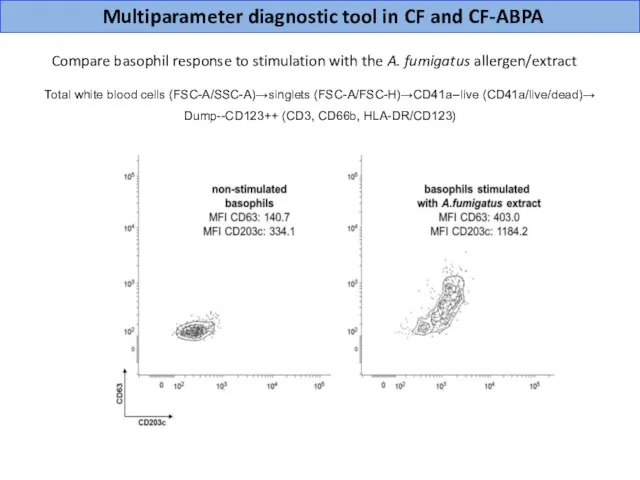
- 25. Multiparameter diagnostic tool in CF and CF-ABPA Compare basophil response to stimulation with the A. fumigatus
- 26. EMD scores based on expression of two independent flow cytometry markers more accurately distinguish allergic (CF-ABPA)
- 27. Cluster matching
- 28. Quantitation of expression levels of surface and intracellular protein biomarkers
- 29. cell cell cell We previously suggested an antigen concentration quantification approach which utilizes the value of
- 30. Experiment: kinetic of mean fluorescence 0.16 min 1 min 3 min 9 min 27 min Beads
- 31. total number of receptor in the volume unit (free+occupied); mean fluorescence value of cell Mathematical model:
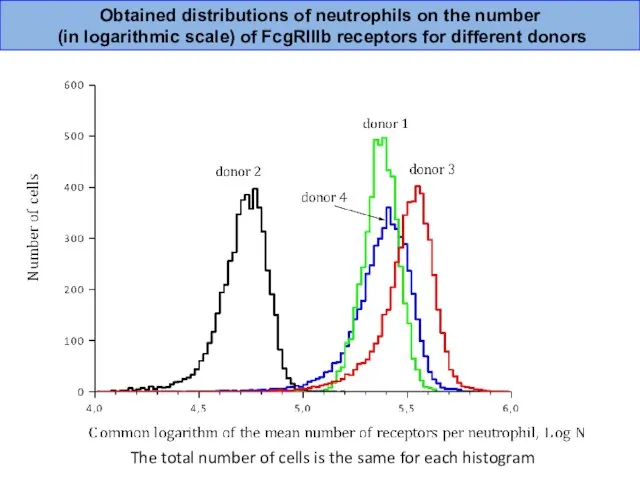
- 32. Obtained distributions of neutrophils on the number (in logarithmic scale) of FcgRIIIb receptors for different donors
- 33. As a solution to this problem we introduce a theoretical approach allowing predicting the binding rate
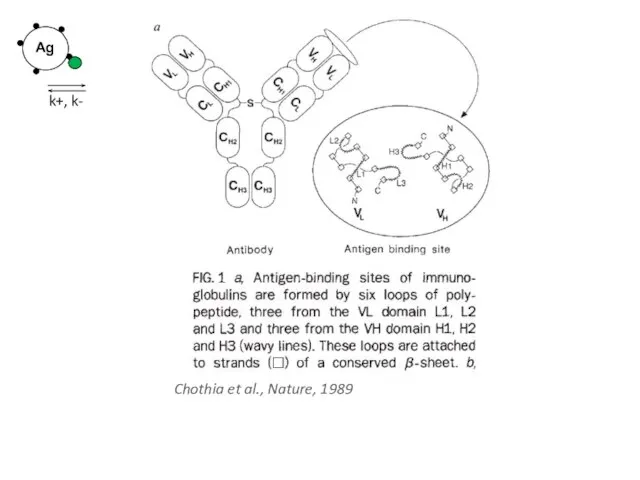
- 34. Chothia et al., Nature, 1989
- 35. Approximation of antigen binding site shape using rectangular “binding spot” model
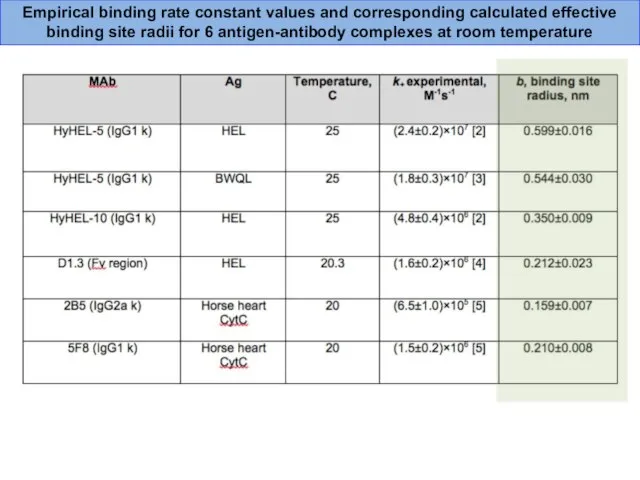
- 36. From the binding rate constant k+, it was possible to estimate the radius b of the
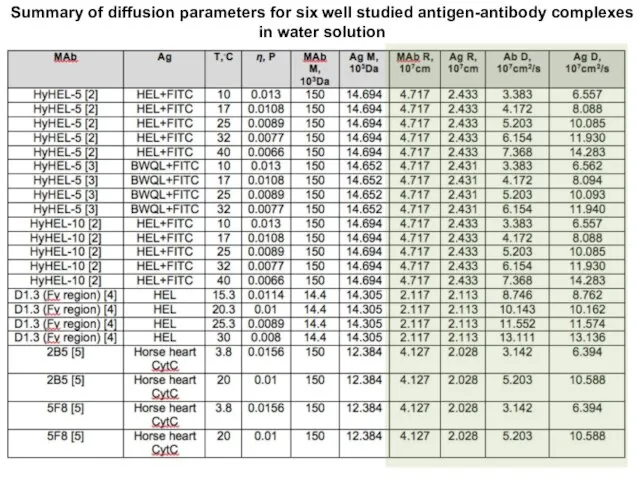
- 37. Empirical binding rate constant values and corresponding calculated effective binding site radii for 6 antigen-antibody complexes
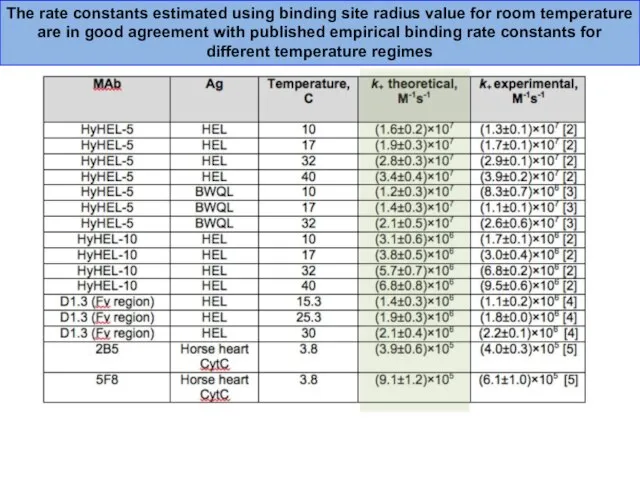
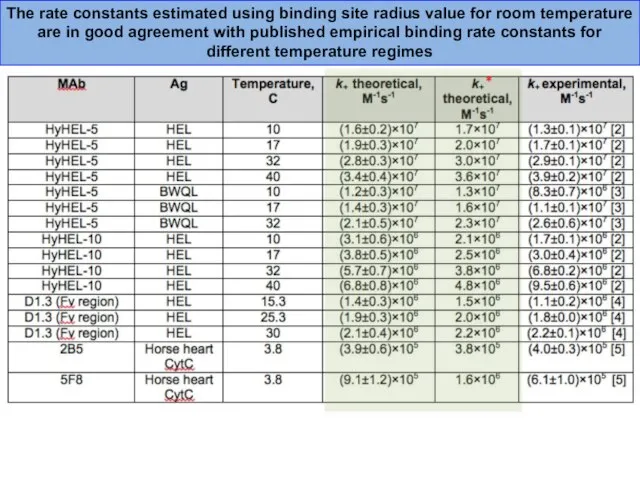
- 38. The rate constants estimated using binding site radius value for room temperature are in good agreement
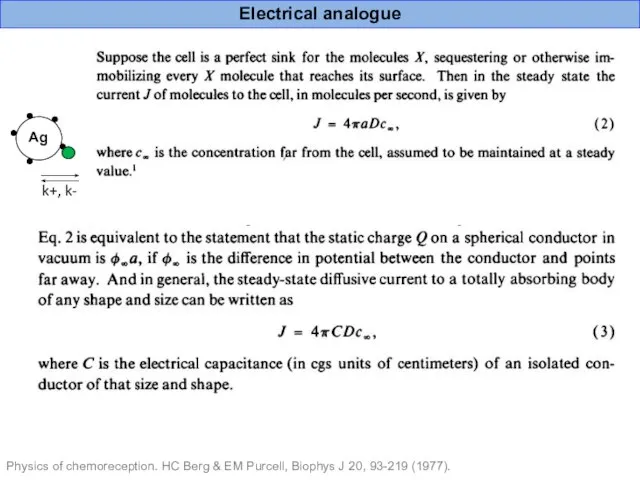
- 39. Physics of chemoreception. HC Berg & EM Purcell, Biophys J 20, 93-219 (1977). Electrical analogue
- 40. Antigen “effective binding site” radius as an equivalent of plate capacitor capacitance “Effective binding site” radius
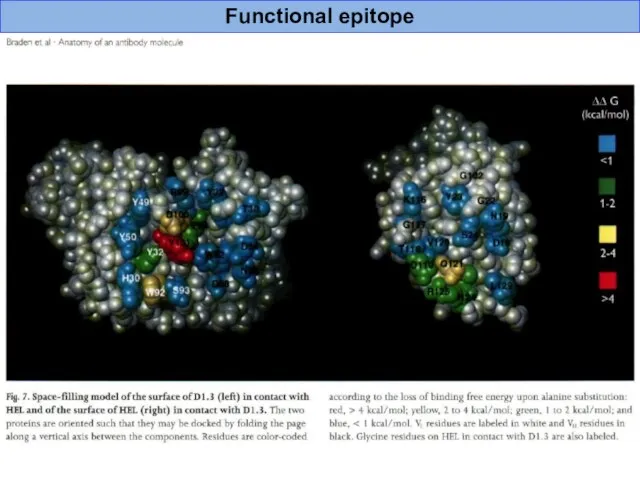
- 41. Functional epitope
- 42. Comparison of estimates for binding site radius electrostatic analogues with effective binding site radii calculated using
- 43. The rate constants estimated using binding site radius value for room temperature are in good agreement
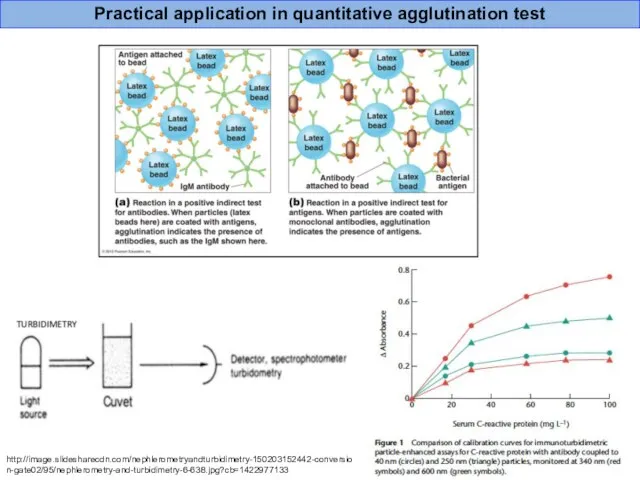
- 44. Practical application in quantitative agglutination test http://image.slidesharecdn.com/nephlerometryandturbidimetry-150203152442-conversion-gate02/95/nephlerometry-and-turbidimetry-6-638.jpg?cb=1422977133
- 45. Thank you! Darya Orlova, Ph.D. Stanford University School of Medicine Genetics Department Beckman Building, Room B013
- 46. [2] Moskalensky et al., 2015 [3] Xavier et al., 1998 [4] Xavier et al., 1999 I5]
- 47. Number of possible 2-D combinations in n dimensional space:
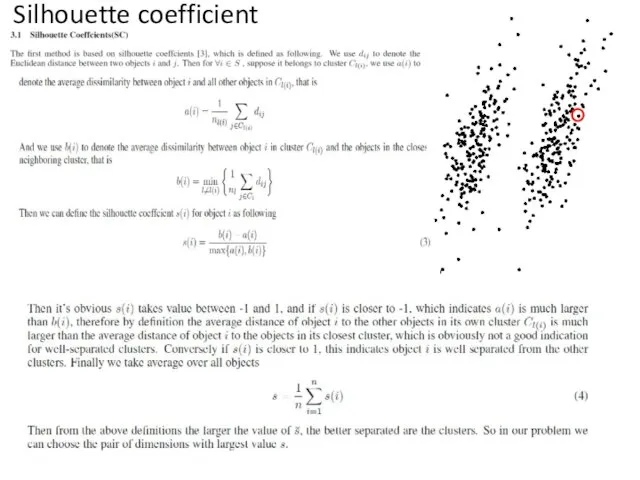
- 51. Silhouette coefficient
- 52. 1. Calculate silhouette coef. (SC) For each pair of clusters+their noise. Calculate % frequency for each
- 53. Summary of diffusion parameters for six well studied antigen-antibody complexes in water solution
- 55. Скачать презентацию


























![[2] Moskalensky et al., 2015 [3] Xavier et al., 1998 [4]](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/510520/slide-45.jpg)




 Презентация на тему Правила поведения в кабинете биологии и химии.
Презентация на тему Правила поведения в кабинете биологии и химии.  Вегетативные органы высших растений. Водоросли
Вегетативные органы высших растений. Водоросли Физиология слуха и равновесия
Физиология слуха и равновесия Наружные покровы человек
Наружные покровы человек Презентация на тему "Моногибридное скрещивание" - скачать презентации по Биологии
Презентация на тему "Моногибридное скрещивание" - скачать презентации по Биологии Строение сердца
Строение сердца Тупорылая Акула
Тупорылая Акула Презентация на тему Направления эволюции урок в 9 классе
Презентация на тему Направления эволюции урок в 9 классе Презентация на тему Движение крови по сосудам
Презентация на тему Движение крови по сосудам Российский Государственный Социальный Университет Факультет Охраны труда и Окружающей среды Кафедра социальной экологии П
Российский Государственный Социальный Университет Факультет Охраны труда и Окружающей среды Кафедра социальной экологии П Углеводный обмен, фотосинтез, гликолиз
Углеводный обмен, фотосинтез, гликолиз Описание эксперимента. Рыба: Данио рерио
Описание эксперимента. Рыба: Данио рерио «Как нерпа приспособилась к жизни в водной среде» Работу выполнила Канахович Александра
«Как нерпа приспособилась к жизни в водной среде» Работу выполнила Канахович Александра Презентация на тему "Подростковые изменения" - скачать бесплатно презентации по Биологии
Презентация на тему "Подростковые изменения" - скачать бесплатно презентации по Биологии Неклеточные формы жизни: В И Р У С Ы
Неклеточные формы жизни: В И Р У С Ы  Полушария большого мозга
Полушария большого мозга Будова вуха, слухового та стато-кінетичного аналізаторів
Будова вуха, слухового та стато-кінетичного аналізаторів Тема: «Опорно-двигательная система». Урок 2. Осевой скелет и скелет конечностей. Перечень слайдов: Строение черепа. Строение чер
Тема: «Опорно-двигательная система». Урок 2. Осевой скелет и скелет конечностей. Перечень слайдов: Строение черепа. Строение чер Птицы тундры. Выполнила: Серых Виктория, 6 класс МКОУ Пензинская ООШ Новосибирская область Барабинский район Руководитель: Серы
Птицы тундры. Выполнила: Серых Виктория, 6 класс МКОУ Пензинская ООШ Новосибирская область Барабинский район Руководитель: Серы Нейропсихология внимания
Нейропсихология внимания Нарушения в работе нервной системы и их предупреждение
Нарушения в работе нервной системы и их предупреждение Еж. Разновидности ежей. Среда обитания
Еж. Разновидности ежей. Среда обитания Гаметогенез, оплодотворение
Гаметогенез, оплодотворение Птицы. Многообразие птичьего мира
Птицы. Многообразие птичьего мира Рефлекс. Рефлекторная дуга
Рефлекс. Рефлекторная дуга Презентация по биологии Взаимодействие аллельных и неаллельных генов.
Презентация по биологии Взаимодействие аллельных и неаллельных генов.  Класс Птицы
Класс Птицы Гистология. Введение в учение о тканях <number>
Гистология. Введение в учение о тканях <number>