Содержание
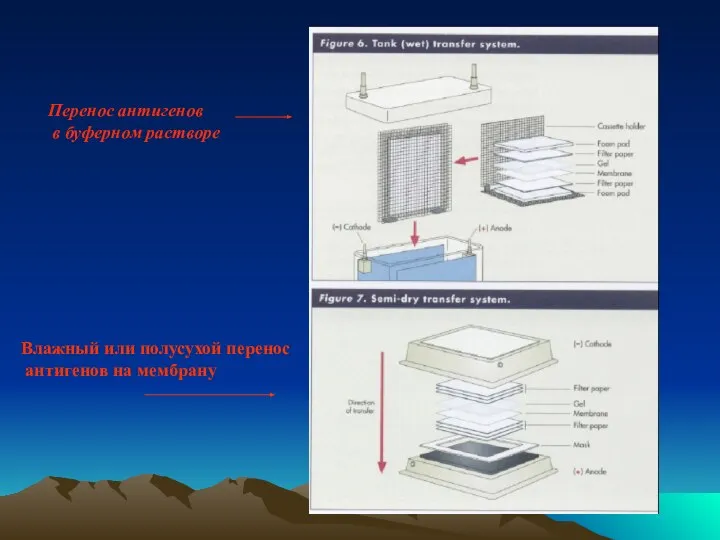
- 32. Перенос антигенов в буферном растворе Влажный или полусухой перенос антигенов на мембрану
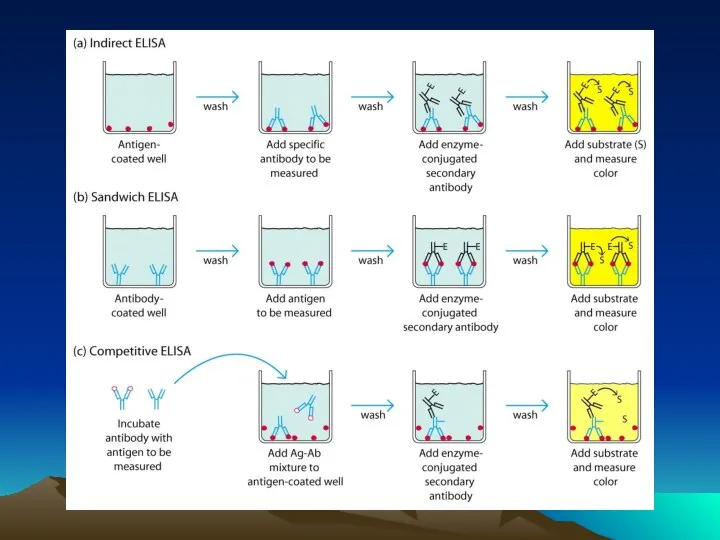
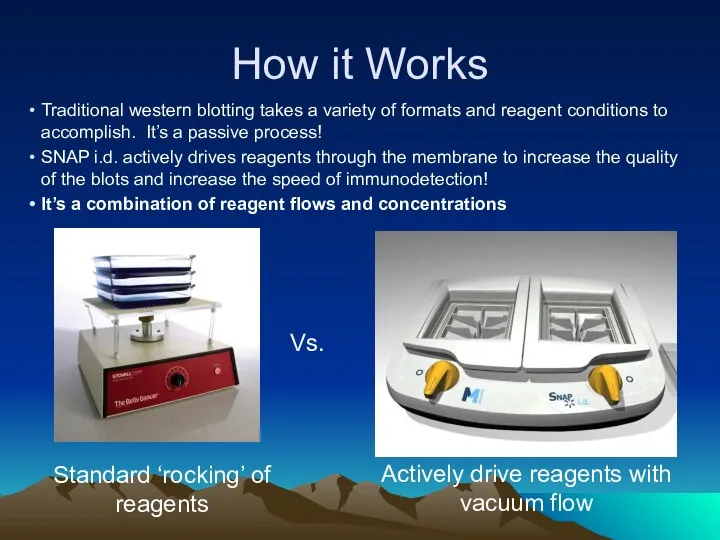
- 33. How it Works Traditional western blotting takes a variety of formats and reagent conditions to accomplish.
- 34. How it Works – reagent flows Gentle Rocking Reagents diffuse slowly into membrane Reagents rapidly driven
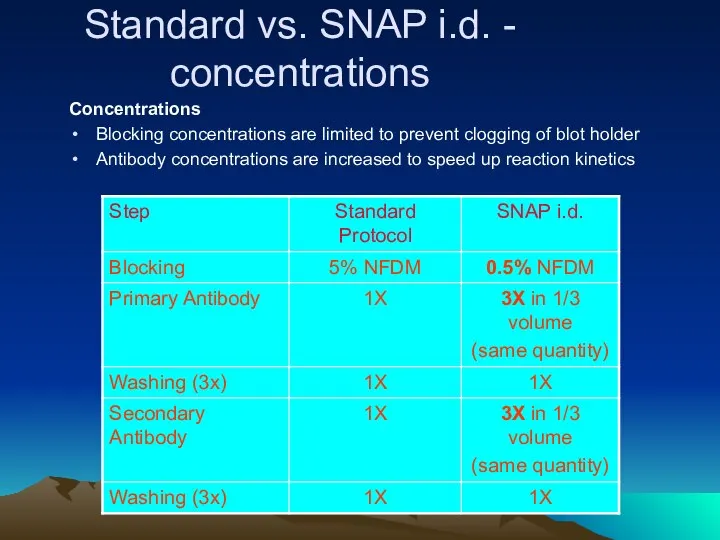
- 35. Standard vs. SNAP i.d. - concentrations Concentrations Blocking concentrations are limited to prevent clogging of blot
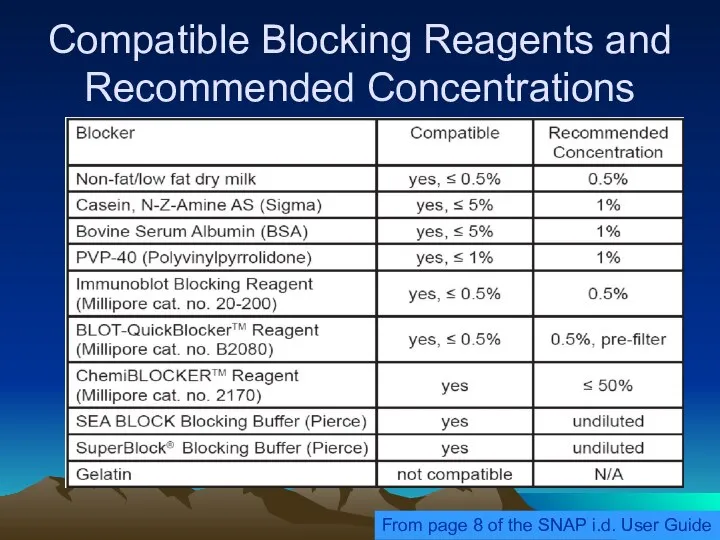
- 36. Compatible Blocking Reagents and Recommended Concentrations From page 8 of the SNAP i.d. User Guide
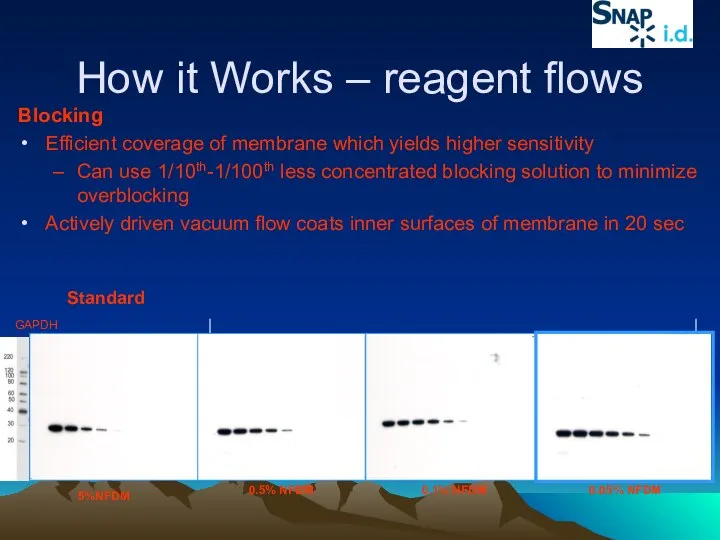
- 37. How it Works – reagent flows Blocking Efficient coverage of membrane which yields higher sensitivity Can
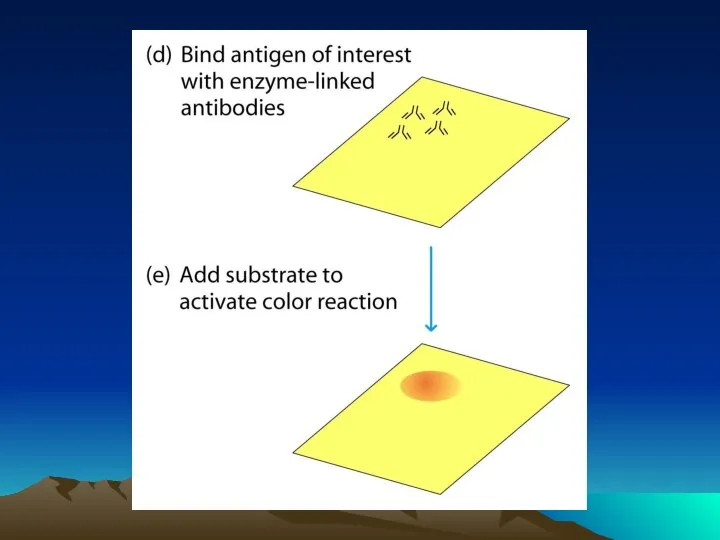
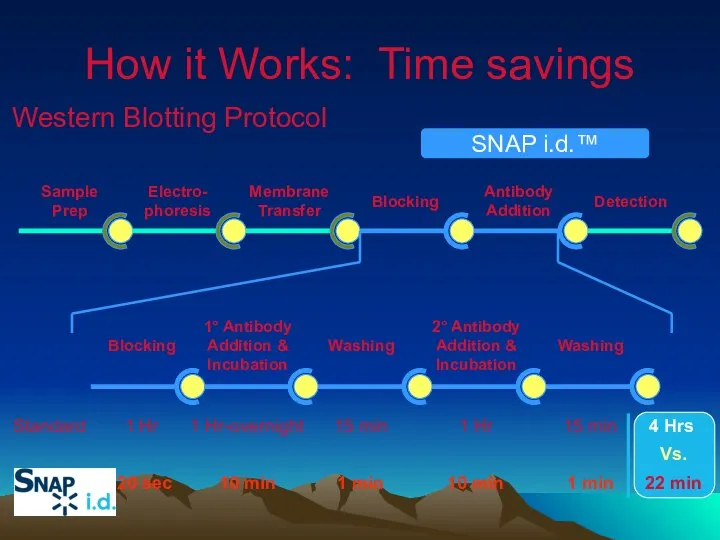
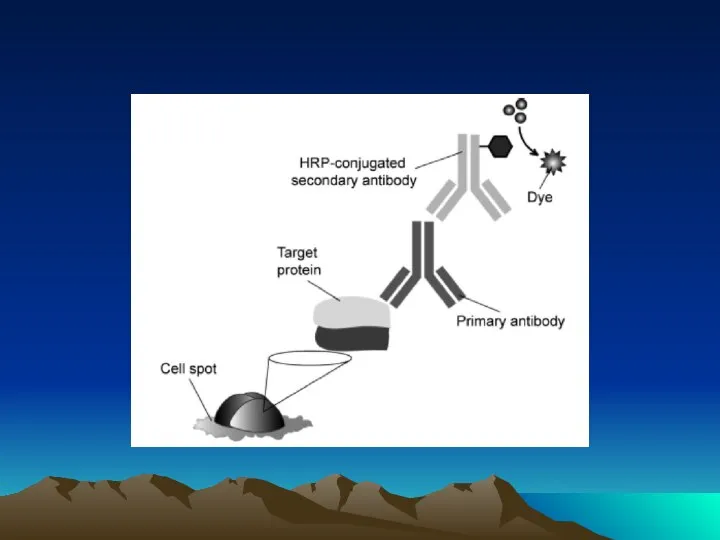
- 38. 1° Antibody Addition & Incubation Washing 2° Antibody Addition & Incubation Washing Blocking 1 Hr 1
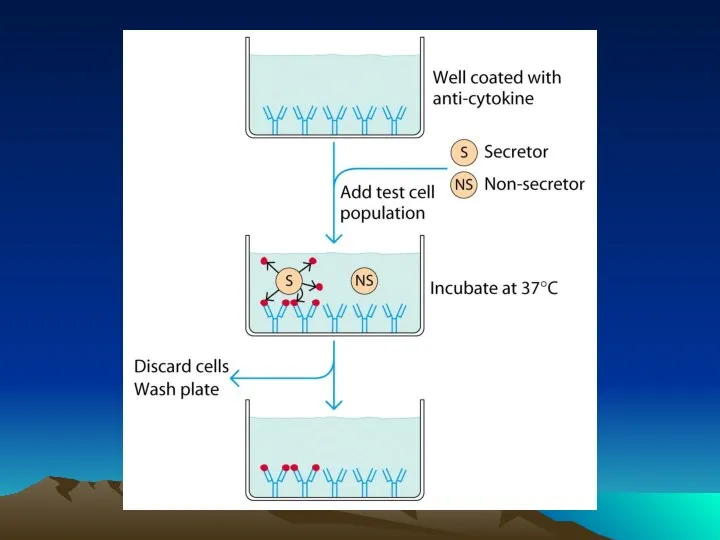
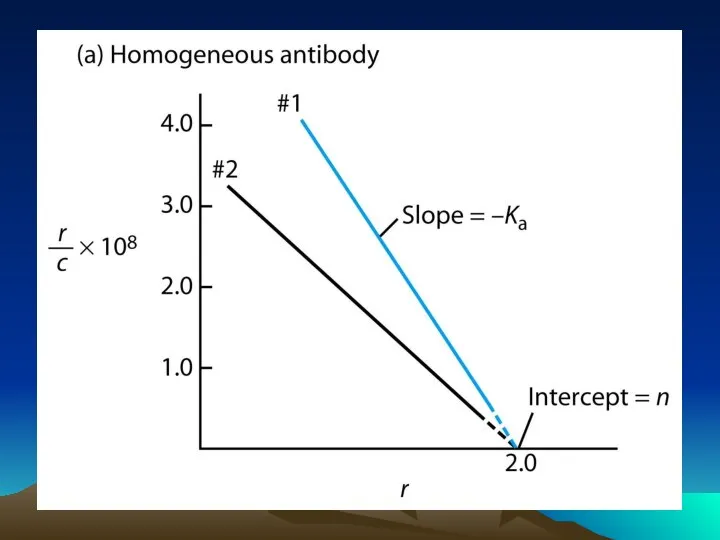
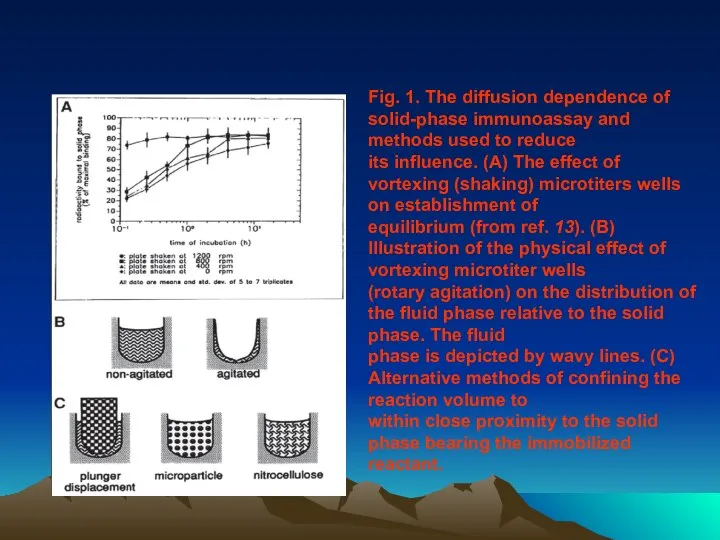
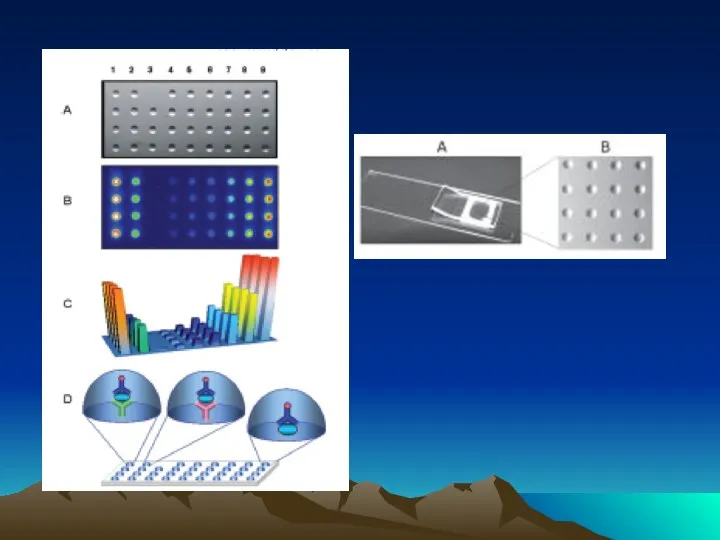
- 42. Fig. 1. The diffusion dependence of solid-phase immunoassay and methods used to reduce its influence. (A)
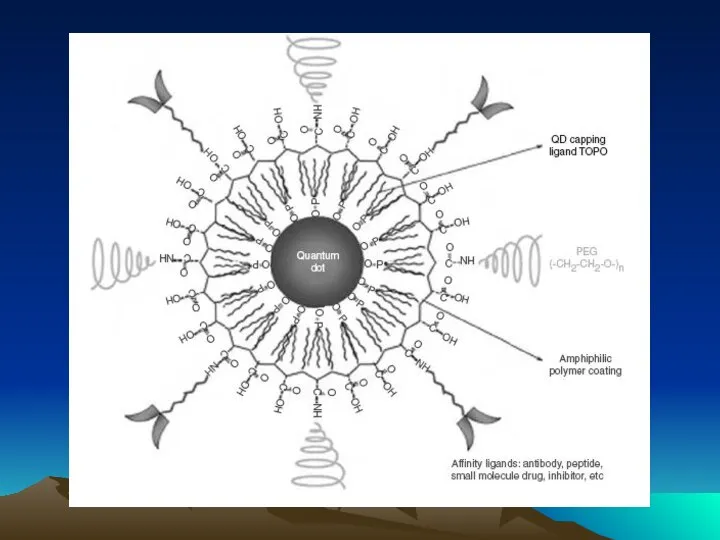
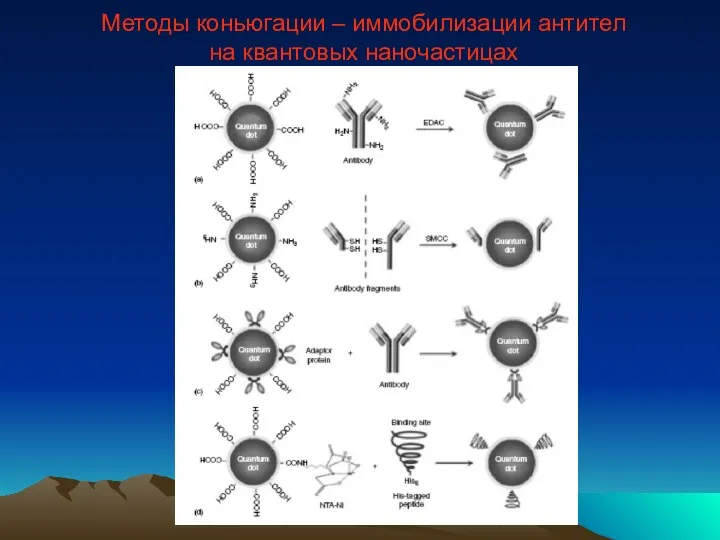
- 49. Методы коньюгации – иммобилизации антител на квантовых наночастицах
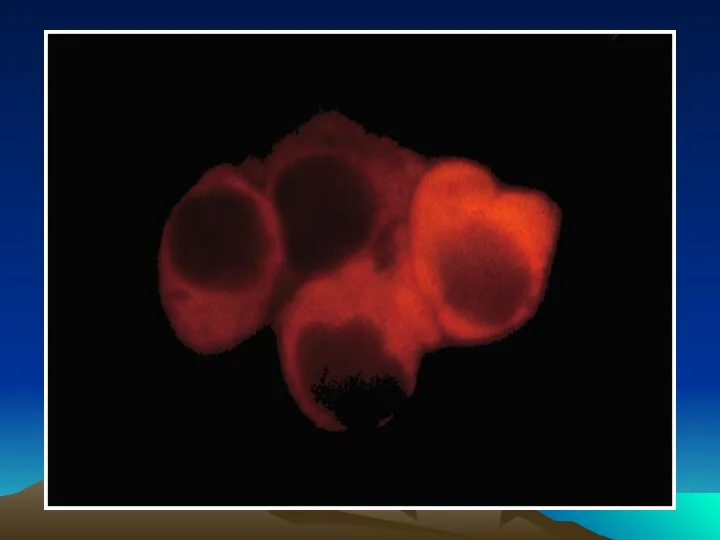
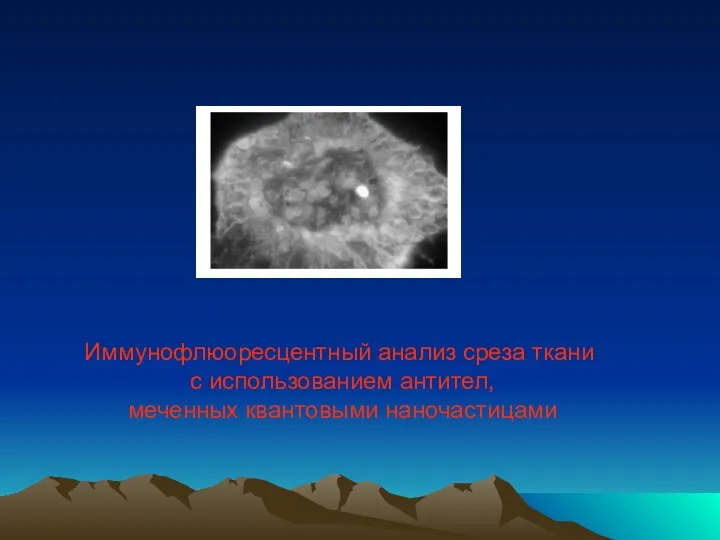
- 50. Иммунофлюоресцентный анализ среза ткани с использованием антител, меченных квантовыми наночастицами
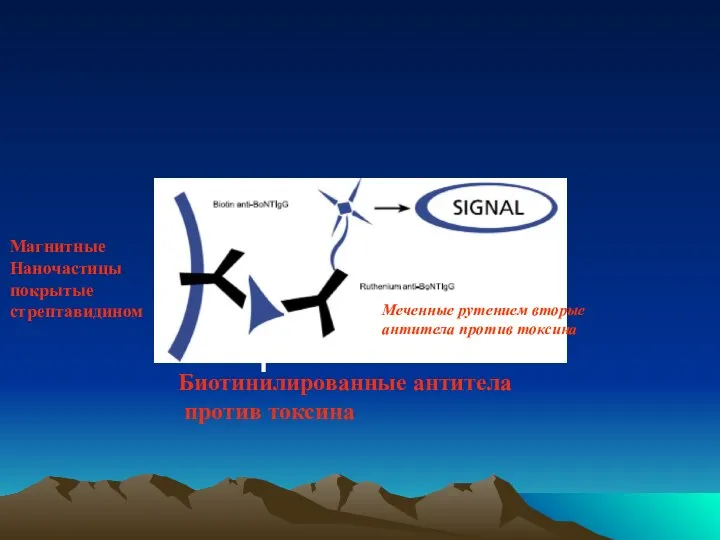
- 53. Магнитные Наночастицы покрытые стрептавидином Биотинилированные антитела против токсина Меченные рутением вторые антитела против токсина
- 54. Чиповая технология с использованием сандвич варианта ИФА и стрептавидин биотиновой ститемы Иммобилизация первых антител на чип,
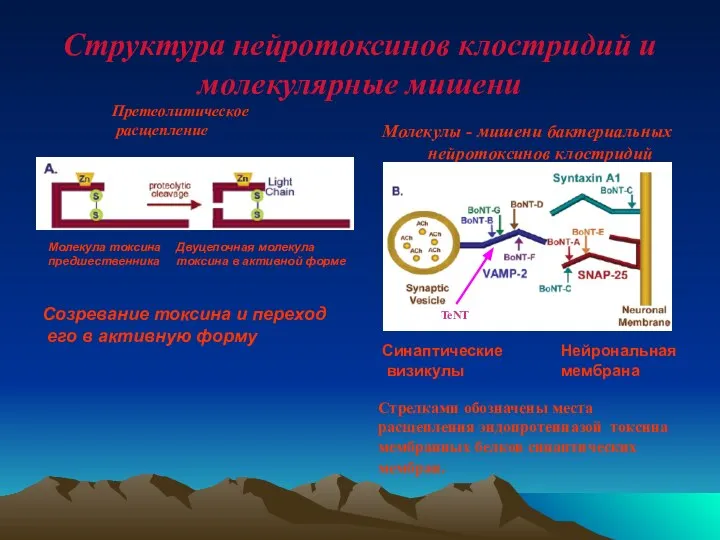
- 56. Структура нейротоксинов клостридий и молекулярные мишени Молекулы - мишени бактериальных нейротоксинов клостридий Молекула токсина предшественника Претеолитическое
- 57. Липосомы-ПЦР иммуноанализ биотоксинов Антитела Определяемый токсин Липосомы, содержащие на поверхности рецептор токсина и фрагменты ДНК внутри
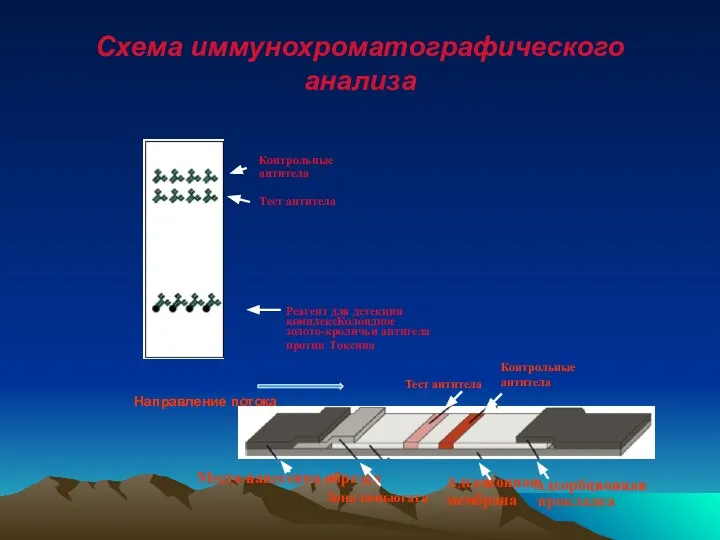
- 58. Схема иммунохроматографического анализа Реагент для детекции комплексКолоидное золото-кроличьи антитела против Токсина Контрольные антитела Тест антитела Тест
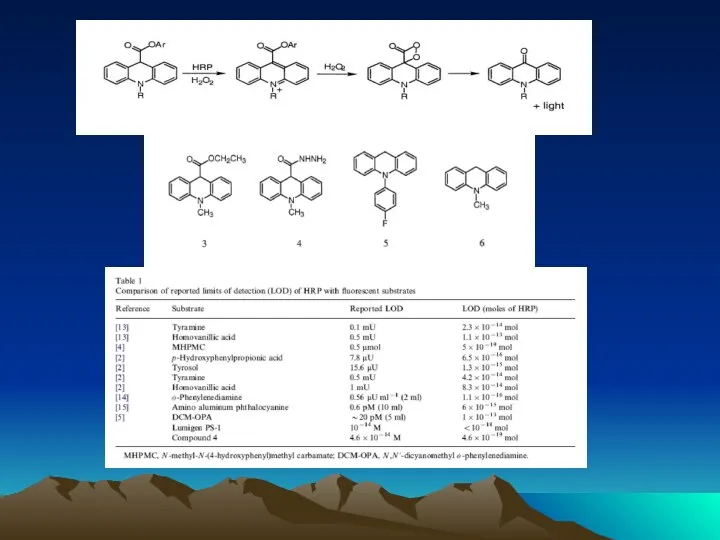
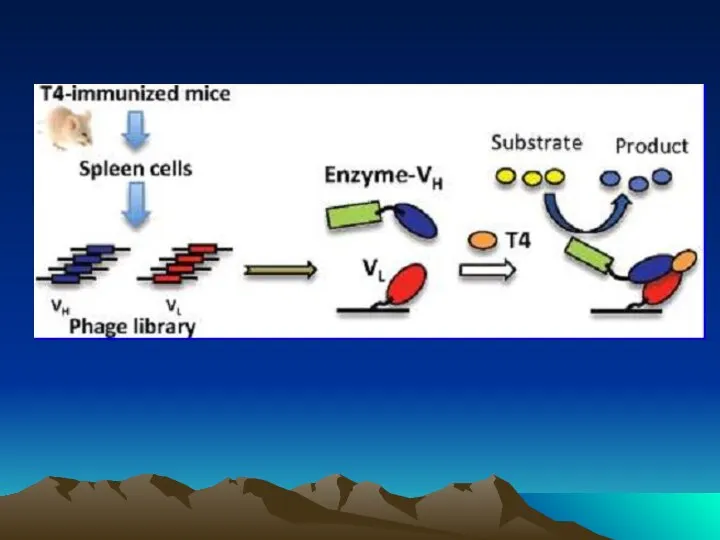
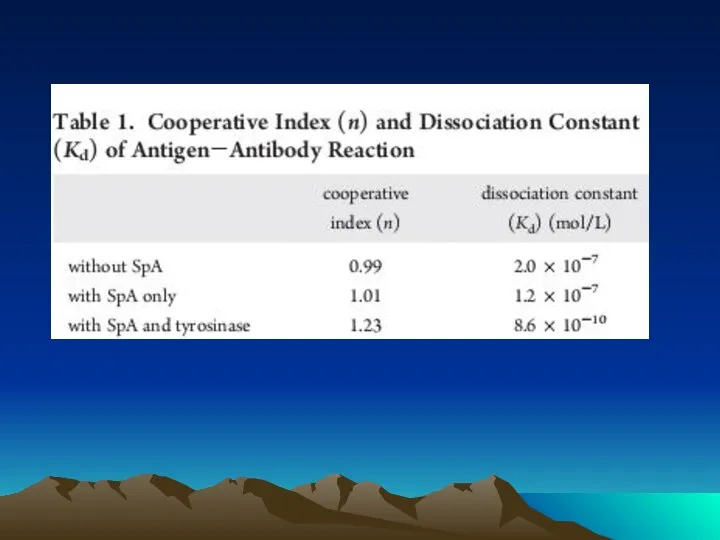
- 72. Conjugated to the amino group carrying platform using tyrosinase. To assess the orientation of immobilized antibodies,
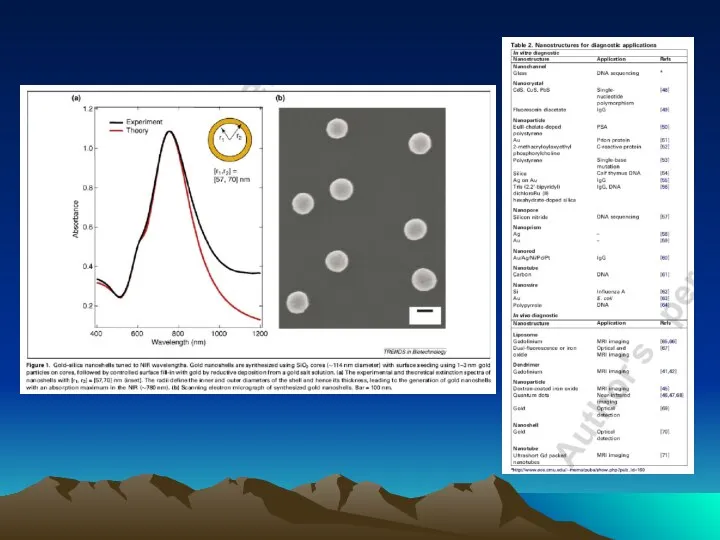
- 77. Representative atomic force microscope images of self-assembled oligomeric DNA–STV conjugates (a) and DNA–STV nanocircles (b). The
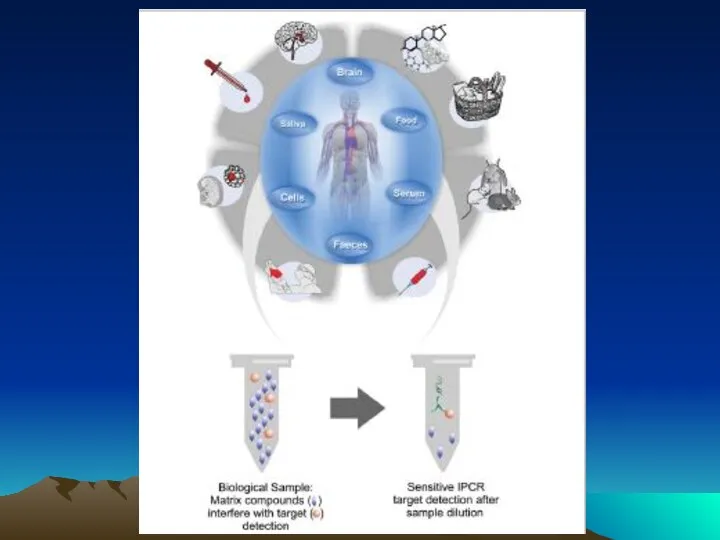
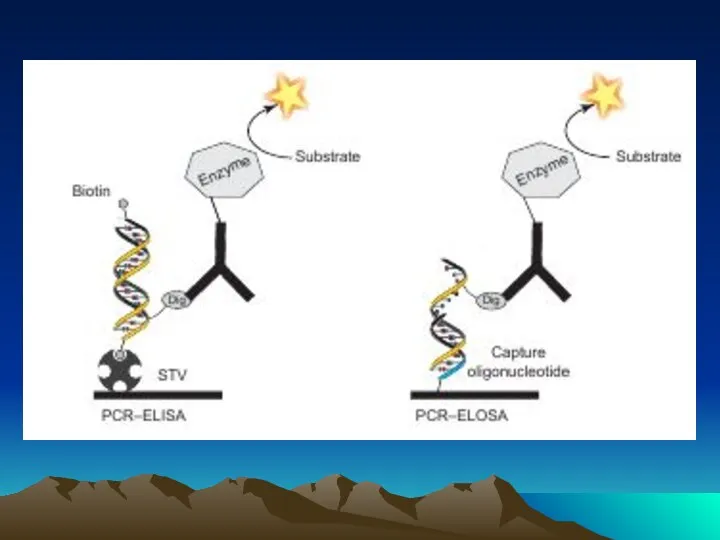
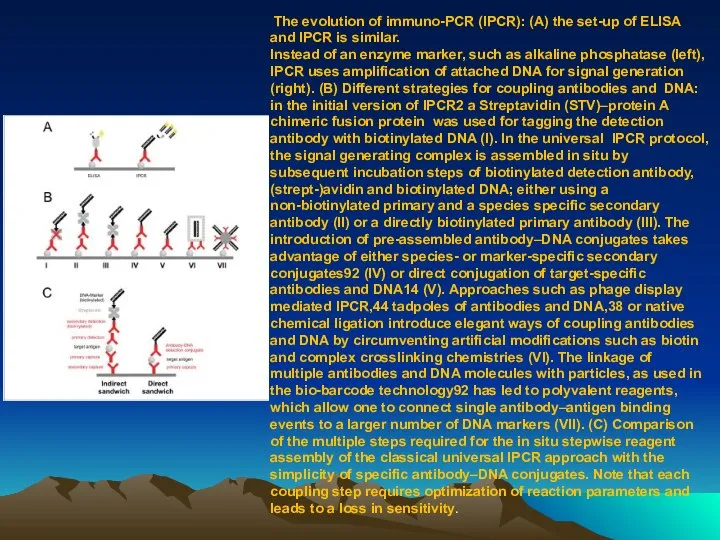
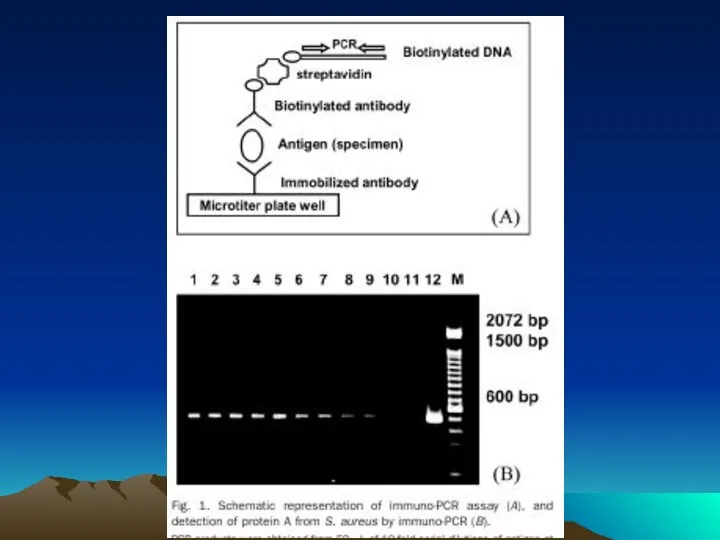
- 78. The evolution of immuno-PCR (IPCR): (A) the set-up of ELISA and IPCR is similar. Instead of
- 79. Typical results of immuno-PCR (IPCR) experiments. (A) Comparison of IPCR, the analogous conventional ELISA for the
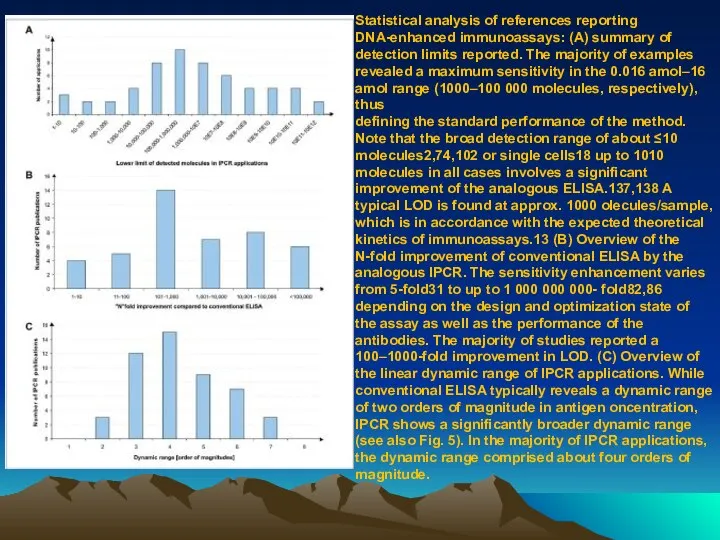
- 80. Statistical analysis of references reporting DNA-enhanced immunoassays: (A) summary of detection limits reported. The majority of
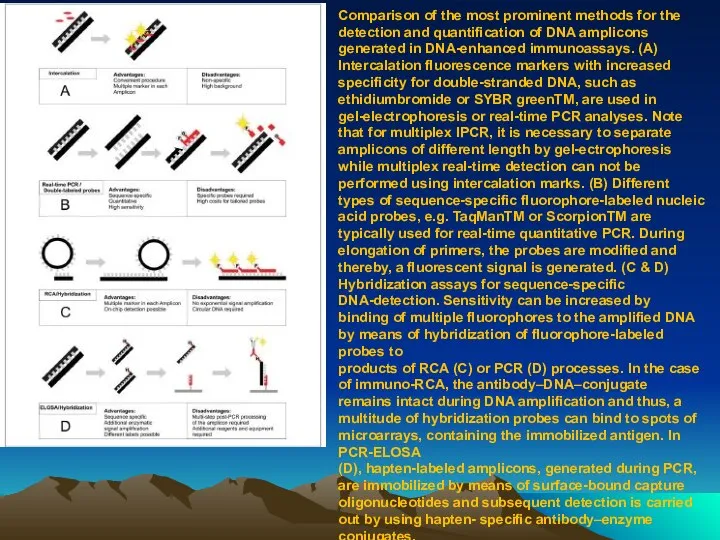
- 81. Comparison of the most prominent methods for the detection and quantification of DNA amplicons generated in
- 86. Multiplex and polyplex assays for the detection of several antigens in a single sample: in multiplex
- 88. Schematic representing the flow of reactions involved in the immuno-PCR signal amplification assay for detection of
- 89. iPCR-SA assay detection of SEA (A) or SEB (B) spiked into tryptic soy broth at select
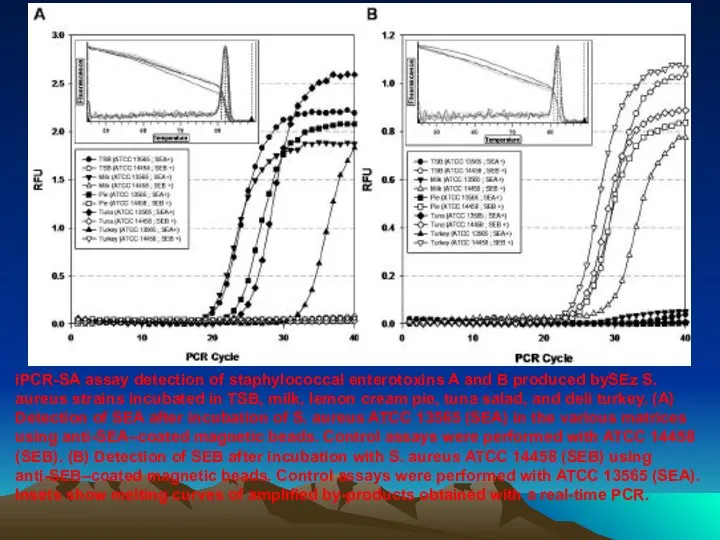
- 90. iPCR-SA assay detection of staphylococcal enterotoxins A and B produced bySEz S. aureus strains incubated in
- 105. Скачать презентацию
























































 Альдегиды и кетоны
Альдегиды и кетоны Машиностроительные материалы
Машиностроительные материалы Изучение свойств пластичных масс для лепки
Изучение свойств пластичных масс для лепки Нанотехнологии. Методы получения наноматериалов. (Лекция 2)
Нанотехнологии. Методы получения наноматериалов. (Лекция 2) Химический диктант как метод формирующего оценивания
Химический диктант как метод формирующего оценивания Тағамдық қоспалардың функционалдық жүктелуі және олардың сипаттамасы
Тағамдық қоспалардың функционалдық жүктелуі және олардың сипаттамасы Сравнительный анализ Фармакопейных статей для субстанций, представленных в мировых Фармакопеях
Сравнительный анализ Фармакопейных статей для субстанций, представленных в мировых Фармакопеях Решение задач на нахождение молекулярной формулы органических веществ
Решение задач на нахождение молекулярной формулы органических веществ Дисахариды и полисахариды
Дисахариды и полисахариды Атомның электрондық конфегурациясы
Атомның электрондық конфегурациясы Скорость химической реакции
Скорость химической реакции Гниение аминокислот в кишечнике
Гниение аминокислот в кишечнике Кристаллохимический анализ типичных структур. Основные характеристики элементарной ячейки
Кристаллохимический анализ типичных структур. Основные характеристики элементарной ячейки Растворы. Термодинамическая теория растворов
Растворы. Термодинамическая теория растворов Элементы группы VII B
Элементы группы VII B Экологический проект Тема: Лабораторное исследование химического состава чипсов Работу выполнила Купавцева Татьяна
Экологический проект Тема: Лабораторное исследование химического состава чипсов Работу выполнила Купавцева Татьяна  Изучение фотокатализа в химии
Изучение фотокатализа в химии Основное уравнение молекулярно-кинетической теории
Основное уравнение молекулярно-кинетической теории Яблочная кладовая Автор работы: Быстрова Анастасия, ученица 9«А» класса Руководитель: Баранова Алевтина Владимировна 2010
Яблочная кладовая Автор работы: Быстрова Анастасия, ученица 9«А» класса Руководитель: Баранова Алевтина Владимировна 2010 Презентация по Химии "Белки" - скачать смотреть _
Презентация по Химии "Белки" - скачать смотреть _ Металлы. Общая характеристика
Металлы. Общая характеристика Своя игра «Знаешь ли ты химические элементы?»
Своя игра «Знаешь ли ты химические элементы?» Свойства металла. Подготовительная группа 2
Свойства металла. Подготовительная группа 2 Адсорбционные равновесия и процессы на подвижных и неподвижных границах раздела фаз
Адсорбционные равновесия и процессы на подвижных и неподвижных границах раздела фаз Закон сохранения массы веществ Омельянчук Т.Е. учитель химии МАОУ ДСОШ №2 г. Домодедово
Закон сохранения массы веществ Омельянчук Т.Е. учитель химии МАОУ ДСОШ №2 г. Домодедово Основы МКТ
Основы МКТ Жири. Жири в природі. Біологічна роль жирів. Жири. Жири в природі. Біологічна роль жирів.
Жири. Жири в природі. Біологічна роль жирів. Жири. Жири в природі. Біологічна роль жирів.  ДИВІНИЛОВИЙ КАУЧУК Підготували Реутенко А., Даниленко А., 11-А
ДИВІНИЛОВИЙ КАУЧУК Підготували Реутенко А., Даниленко А., 11-А