Содержание
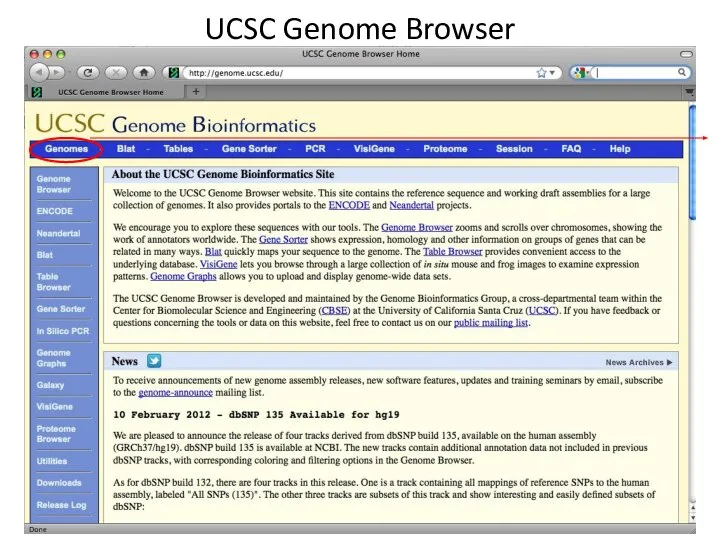
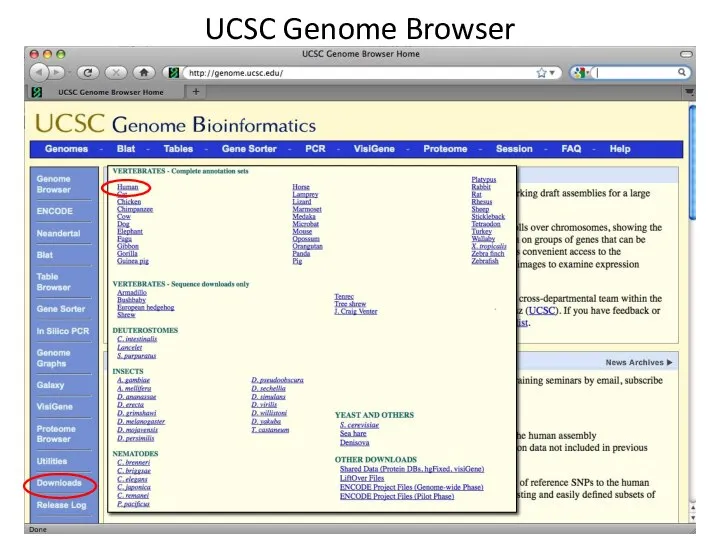
- 2. UCSC Genome Browser
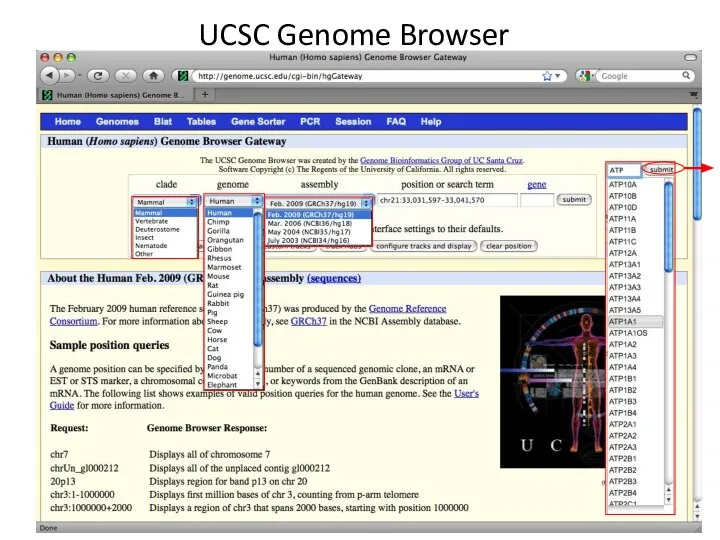
- 3. UCSC Genome Browser
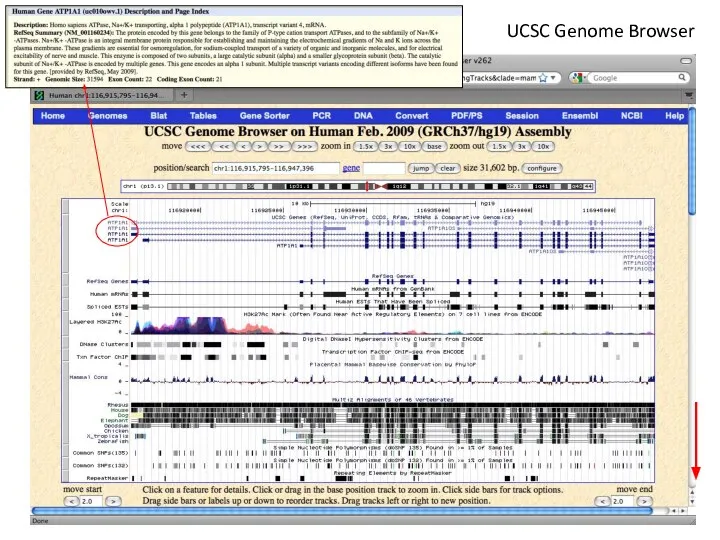
- 4. UCSC Genome Browser
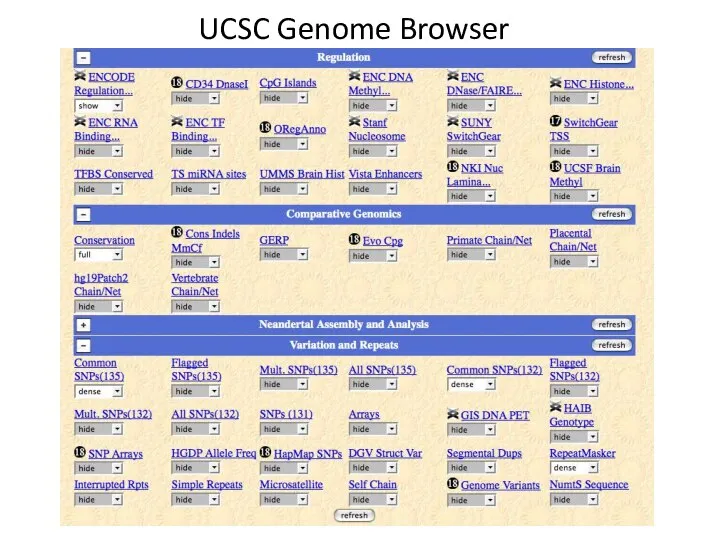
- 5. UCSC Genome Browser
- 6. UCSC Genome Browser
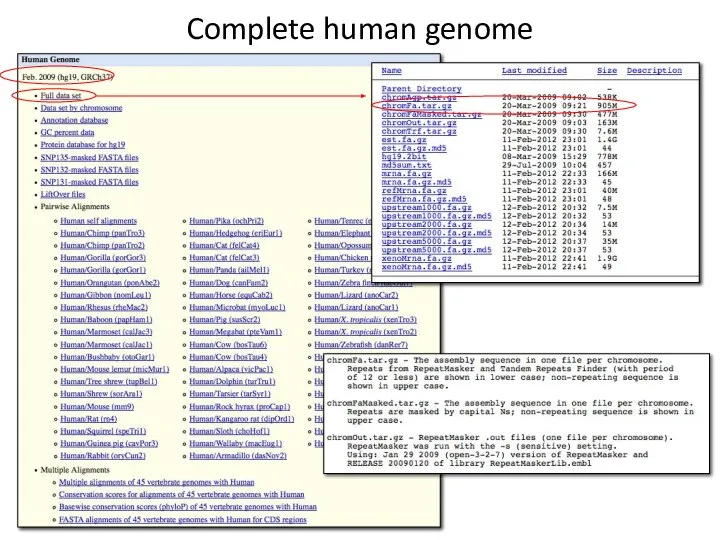
- 7. Complete human genome
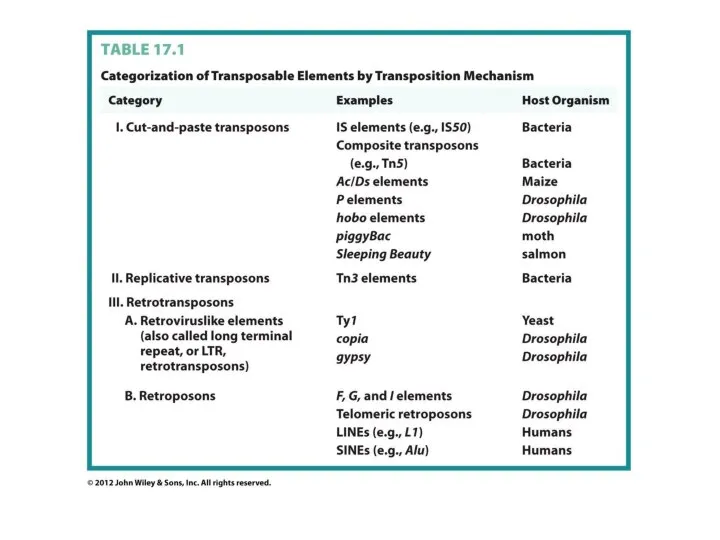
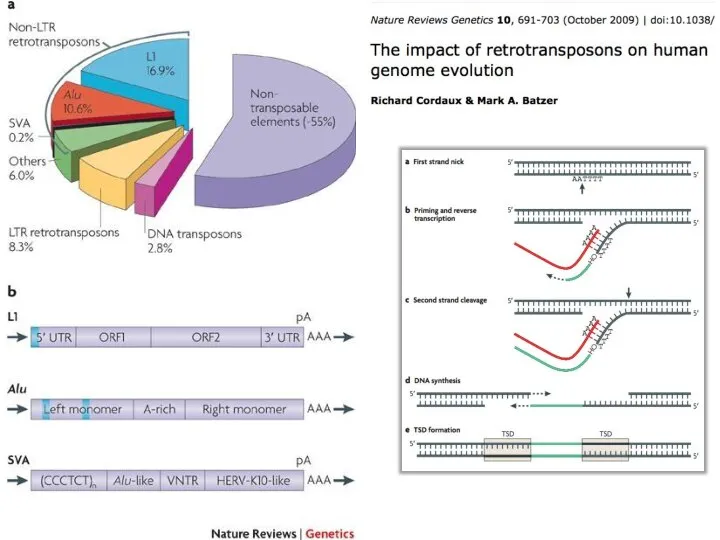
- 8. Transposable Elements 45% of the human genome is occupied by transposons and transposon-like repetitive elements. Barbara
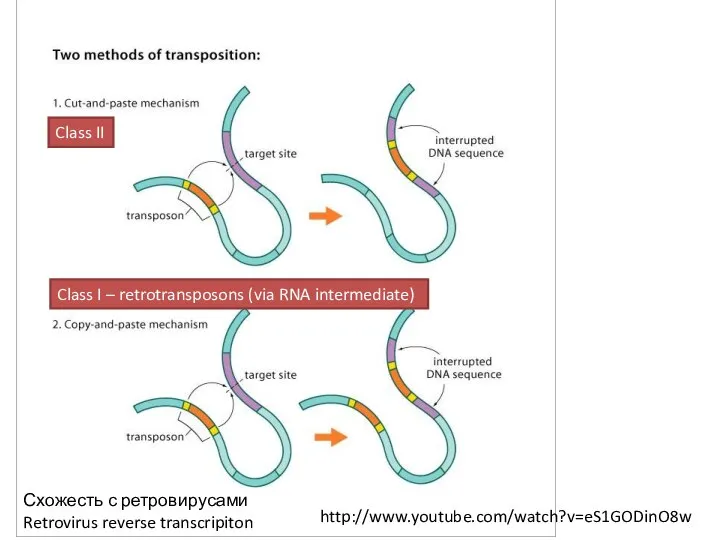
- 9. Схожесть с ретровирусами Retrovirus reverse transcripiton http://www.youtube.com/watch?v=eS1GODinO8w Class II Class I – retrotransposons (via RNA intermediate)
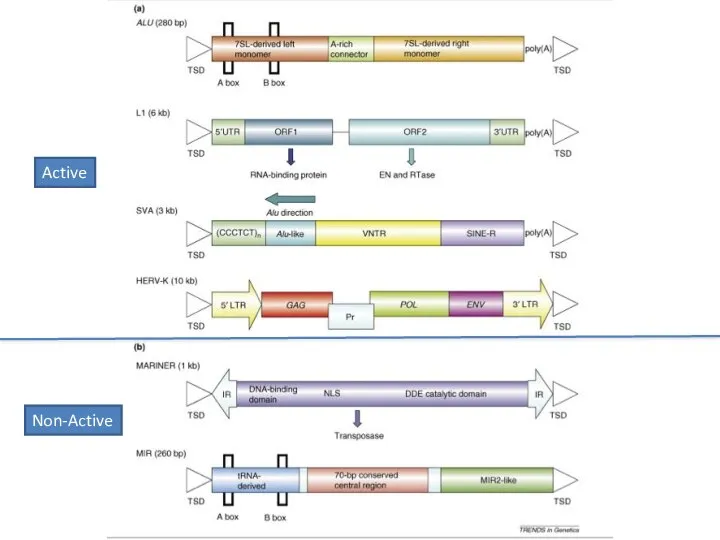
- 13. Active Non-Active
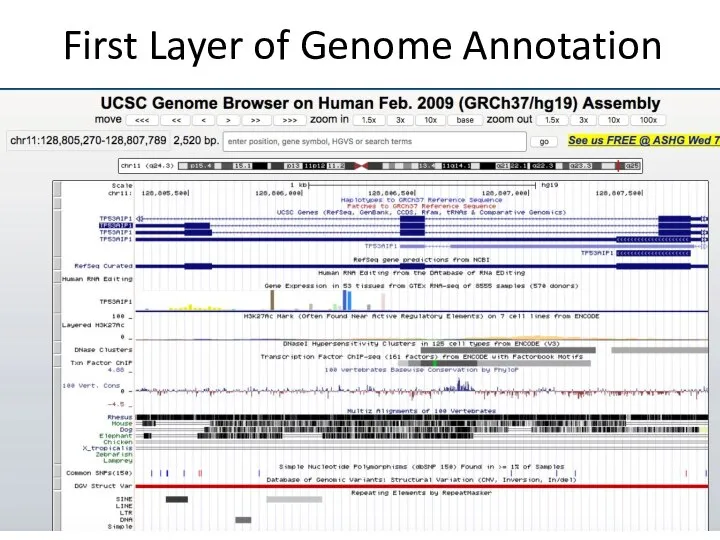
- 14. First Layer of Genome Annotation
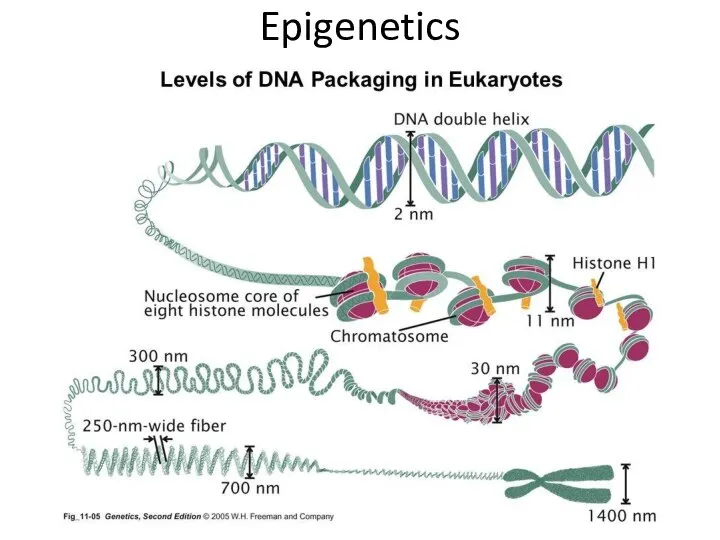
- 15. Epigenetics
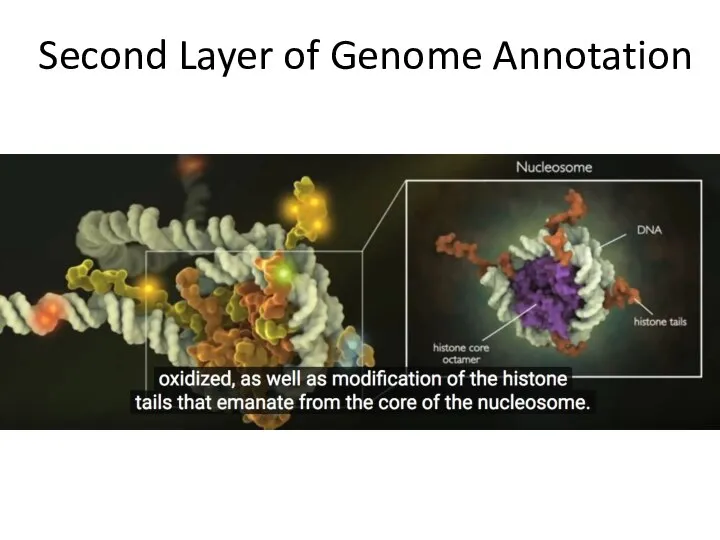
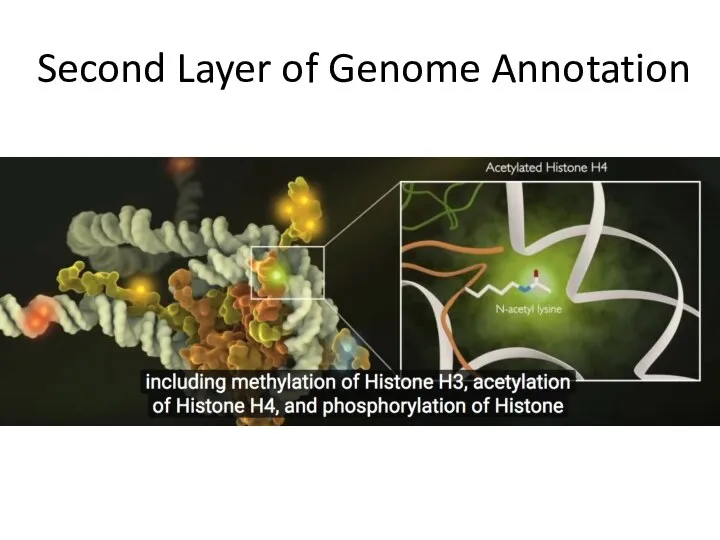
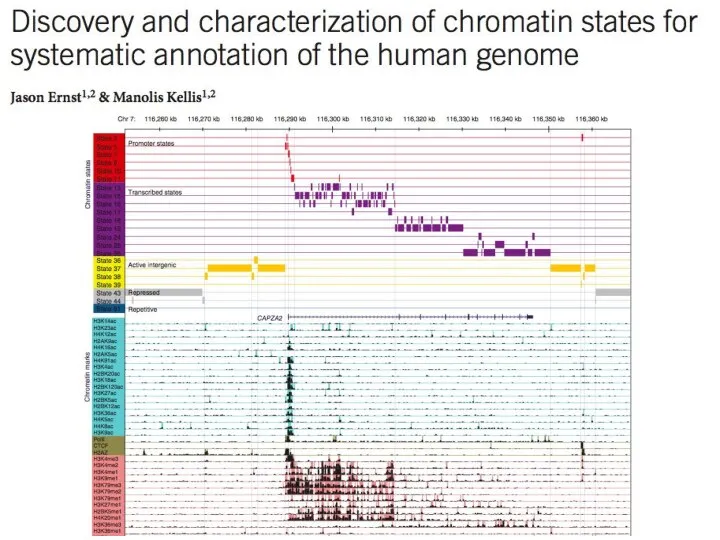
- 16. Second Layer of Genome Annotation
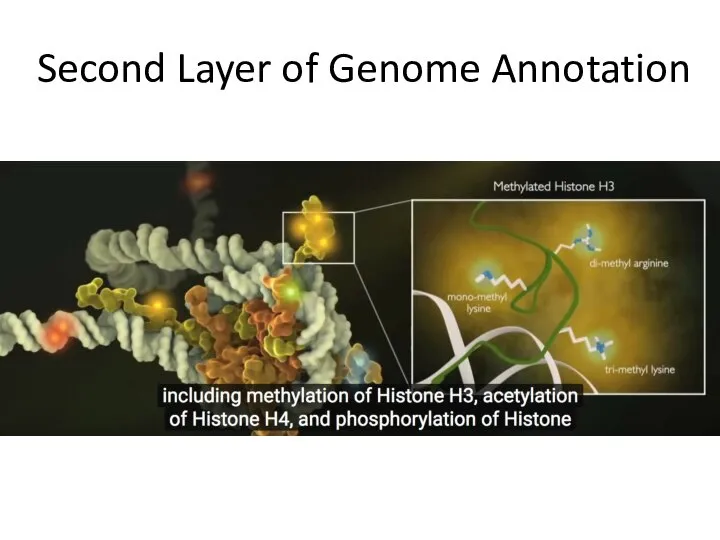
- 17. Second Layer of Genome Annotation
- 18. Second Layer of Genome Annotation
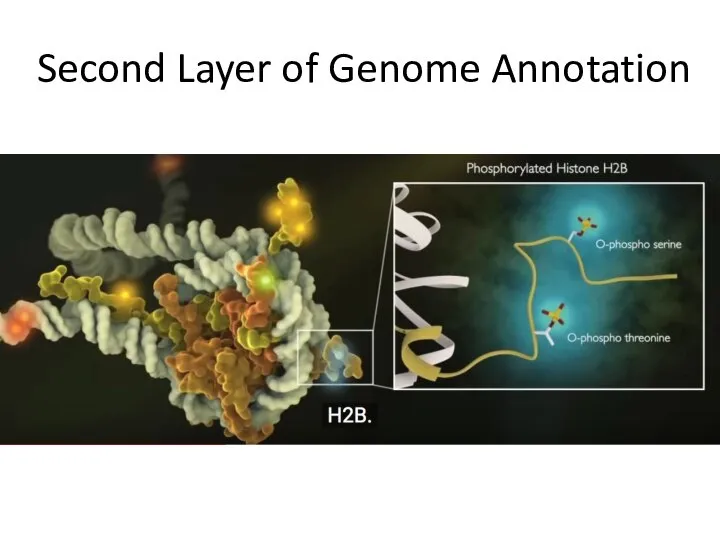
- 19. Second Layer of Genome Annotation
- 20. Second Layer of Genome Annotation
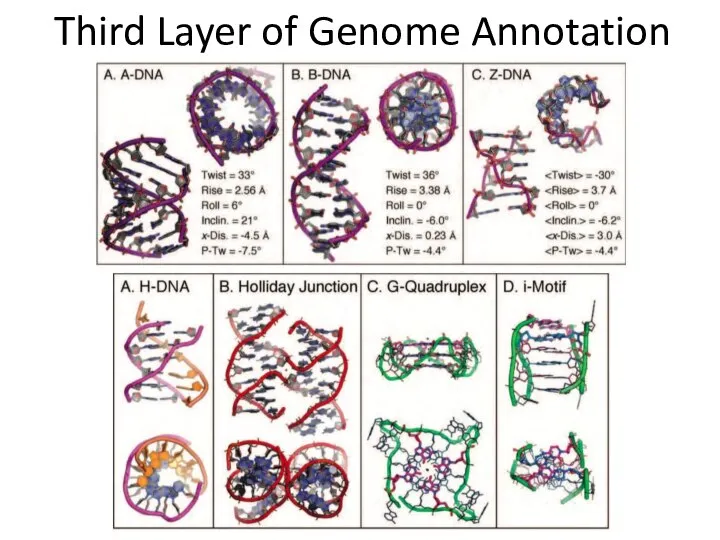
- 21. Third Layer of Genome Annotation
- 22. Data Accumulation
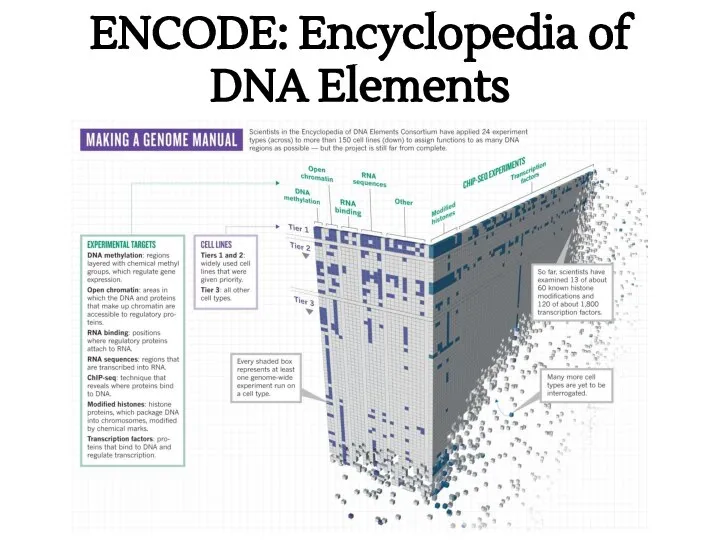
- 23. ENCODE: Encyclopedia of DNA Elements
- 24. Digital Universe Like the Physical Universe the Digital Universe is also expanding but much faster doubling
- 25. Digital Universe Data Universe Will Expand To 44 Trillion GBs By 2020
- 26. Что делать?
- 27. Что получилось? (Success Stories)
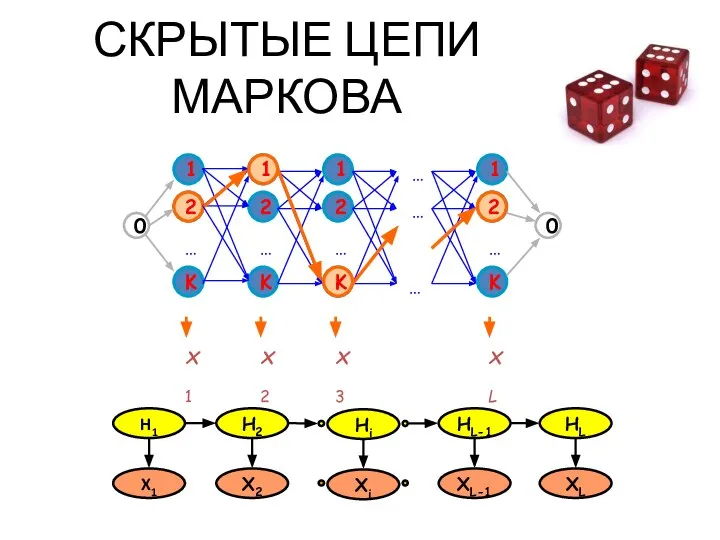
- 28. СКРЫТЫЕ ЦЕПИ МАРКОВА
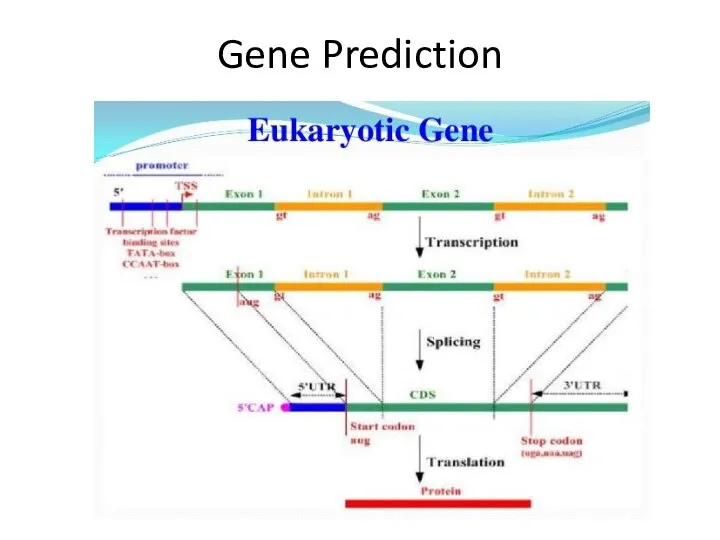
- 29. Gene Prediction
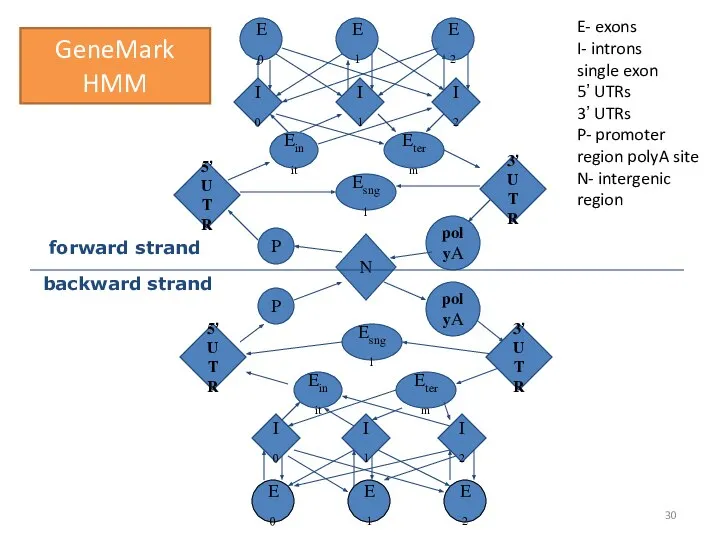
- 30. E0 E1 E2 E2 E1 E0 N P Eterm P Einit polyA 5’ UTR I0 I1
- 31. Promoter prediction Hidden Markov model with six interpolated Markov chain submodels upstream 1 and 2, TATA
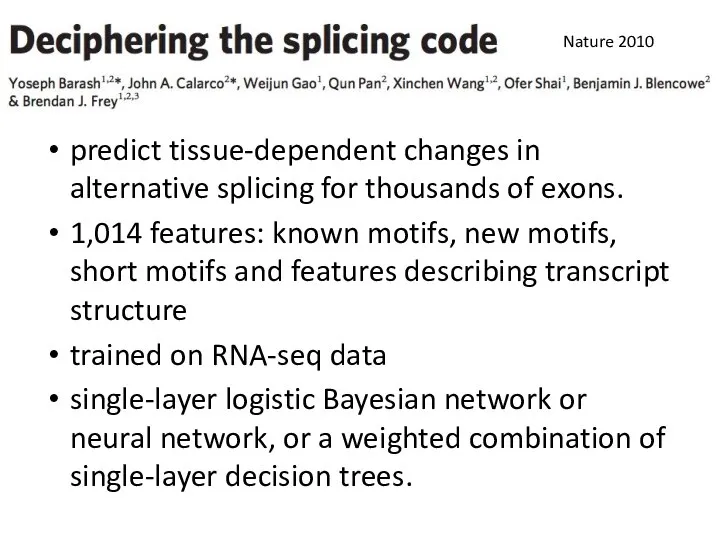
- 32. predict tissue-dependent changes in alternative splicing for thousands of exons. 1,014 features: known motifs, new motifs,
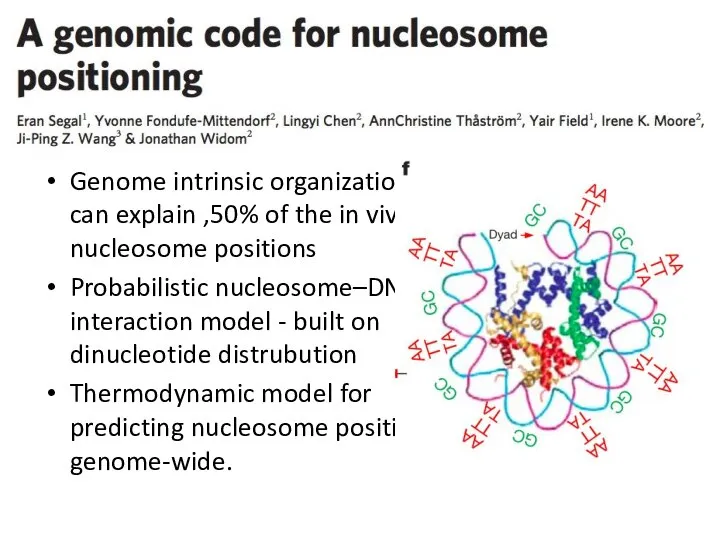
- 33. Genome intrinsic organization can explain ,50% of the in vivo nucleosome positions Probabilistic nucleosome–DNA interaction model
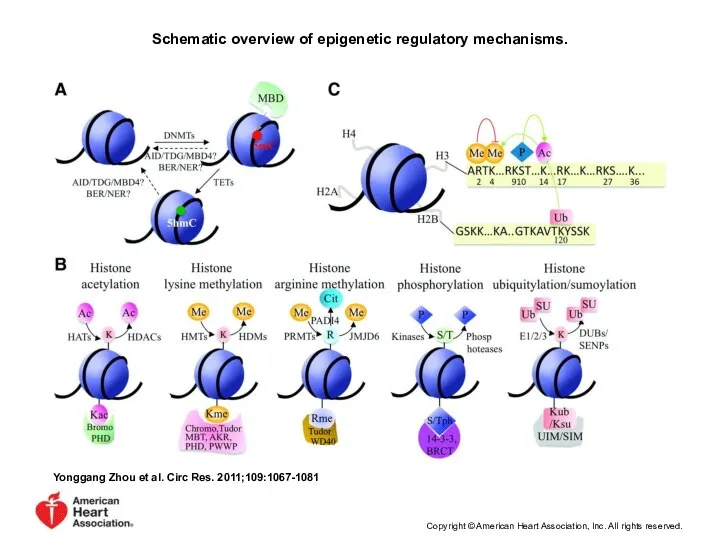
- 35. Schematic overview of epigenetic regulatory mechanisms. Yonggang Zhou et al. Circ Res. 2011;109:1067-1081 Copyright © American
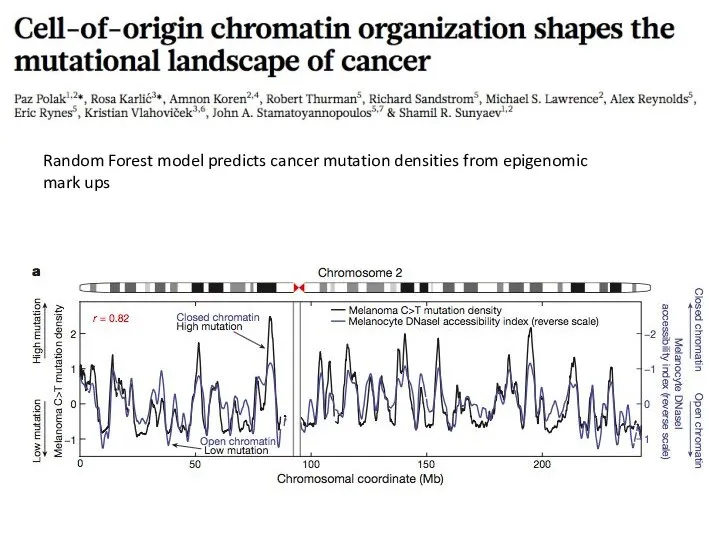
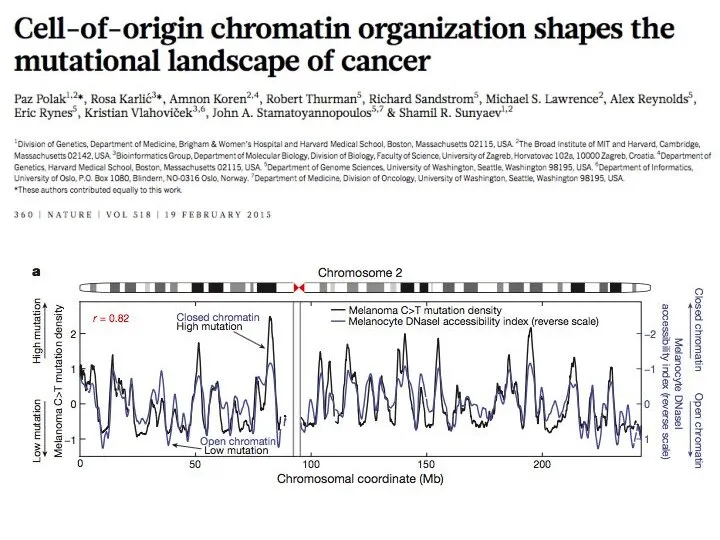
- 36. Random Forest model predicts cancer mutation densities from epigenomic mark ups
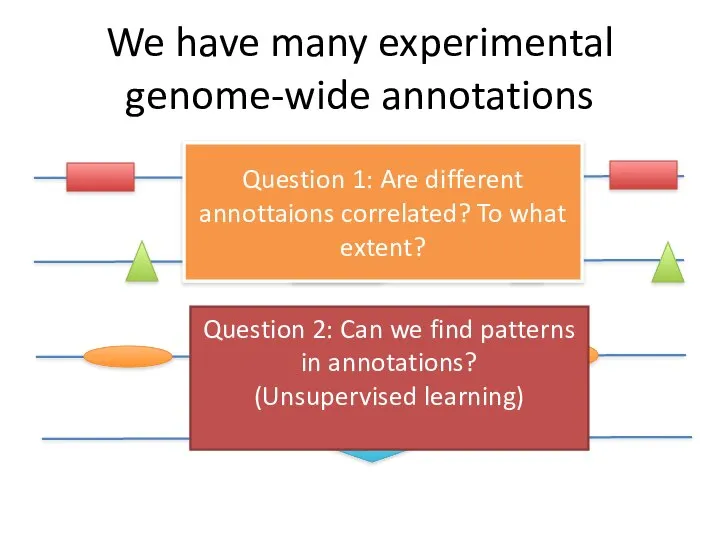
- 38. We have many experimental genome-wide annotations Question 1: Are different annottaions correlated? To what extent? Question
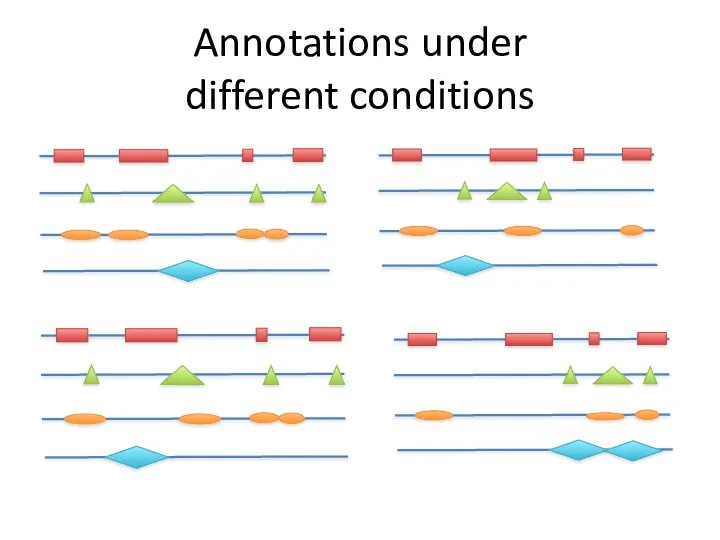
- 39. Annotations under different conditions
- 40. Как много данных? Roadmap Epigenomics ~ 3 000 полногеномных данных ENCODE Encyclopedia of Genomic Elements ~
- 41. Открытые вопросы Какие участки кода работают одновременно? Как переключать режимы работы клетки? Как перепрограммируется код для
- 43. Скачать презентацию








 Средний отдел пищеварительной системы. Гистология
Средний отдел пищеварительной системы. Гистология Механизмы сна. Физиологические изменения во время сна
Механизмы сна. Физиологические изменения во время сна Технологии и оборудование для производства твердых лекарственных форм
Технологии и оборудование для производства твердых лекарственных форм Эритропоэздің нейро-гуморальдық реттелуі
Эритропоэздің нейро-гуморальдық реттелуі Экспериментальная хирургия легких
Экспериментальная хирургия легких Природные лекарственные средства
Природные лекарственные средства Искусственный интеллект. Диагностика в клинике. Визуализационные исследования
Искусственный интеллект. Диагностика в клинике. Визуализационные исследования Бронхиальная астма у детей. Этиология, патогенез, клиника, диагностика, дифференциальный диагноз
Бронхиальная астма у детей. Этиология, патогенез, клиника, диагностика, дифференциальный диагноз Лазерное лечение глаукомы и катаракты
Лазерное лечение глаукомы и катаракты Частная миология Классификация и топография мышц туловища: грудь, спина, живот
Частная миология Классификация и топография мышц туловища: грудь, спина, живот Посттравматический остеомиелит
Посттравматический остеомиелит Синдром бронхообструкции. Дифференцированное применение бронхолитиков
Синдром бронхообструкции. Дифференцированное применение бронхолитиков Введение в венерологию. Современная эпидемиология венерических болезней. Общее течение сифилиса
Введение в венерологию. Современная эпидемиология венерических болезней. Общее течение сифилиса Jmed. Система управления клиникой
Jmed. Система управления клиникой АЧС. Медецина
АЧС. Медецина Жиры с точки зрения ЗОЖ
Жиры с точки зрения ЗОЖ Онтофилогенетические пороки развития кровеносной системы
Онтофилогенетические пороки развития кровеносной системы РОО Врачи Санкт-Петербурга. Повышение качества медицинской помощи
РОО Врачи Санкт-Петербурга. Повышение качества медицинской помощи Классификация вредных привычек. Роль вредных привычек у детей при формировании зубо-челюстных аномалий
Классификация вредных привычек. Роль вредных привычек у детей при формировании зубо-челюстных аномалий Статистические и спектральные показатели ритмокардиоинтервалографии
Статистические и спектральные показатели ритмокардиоинтервалографии Этиологическая диагностика внебольничных инфекций дыхательных путей
Этиологическая диагностика внебольничных инфекций дыхательных путей Су–Джок терапия
Су–Джок терапия Дуглас Дж. Ри Глаукома
Дуглас Дж. Ри Глаукома Инфаркт миокарда
Инфаркт миокарда Анатомо-физиологические особенности детского организма с позиции анестезиолога
Анатомо-физиологические особенности детского организма с позиции анестезиолога Лейкемия. Типы лейкемий
Лейкемия. Типы лейкемий Мимика и жесты в деловом общении
Мимика и жесты в деловом общении Школа здоровья для беременных. Занятие 5. Роды
Школа здоровья для беременных. Занятие 5. Роды