Содержание
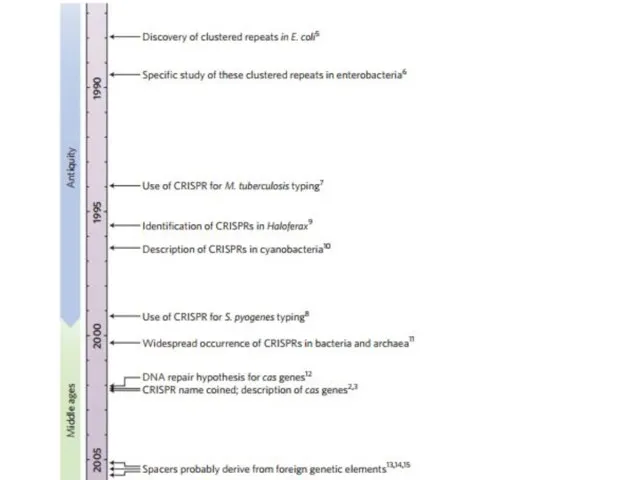
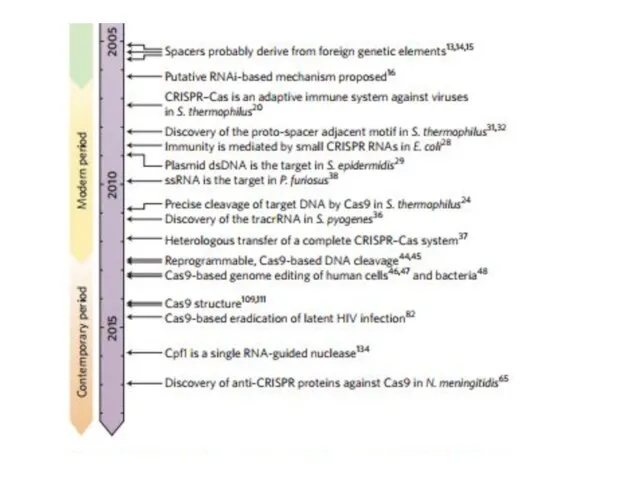
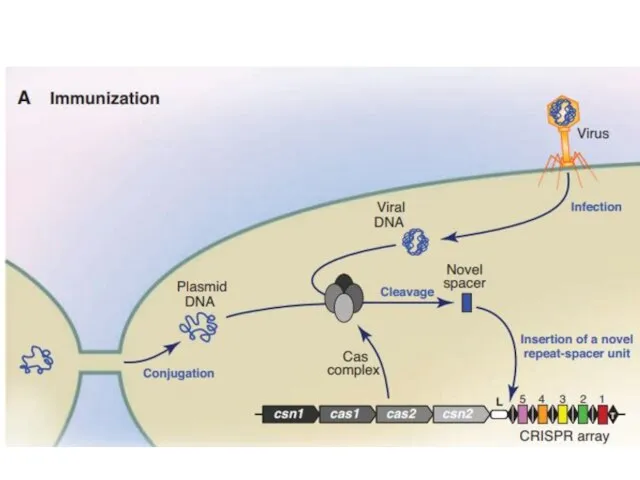
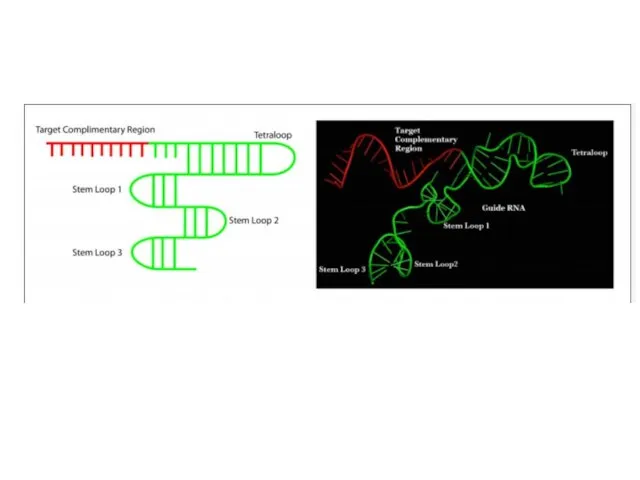
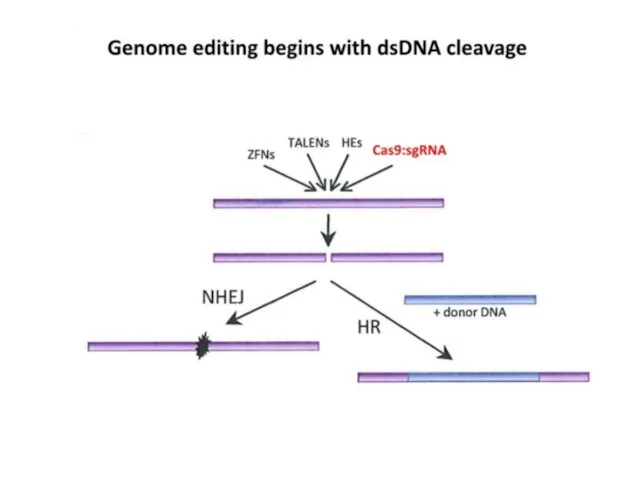
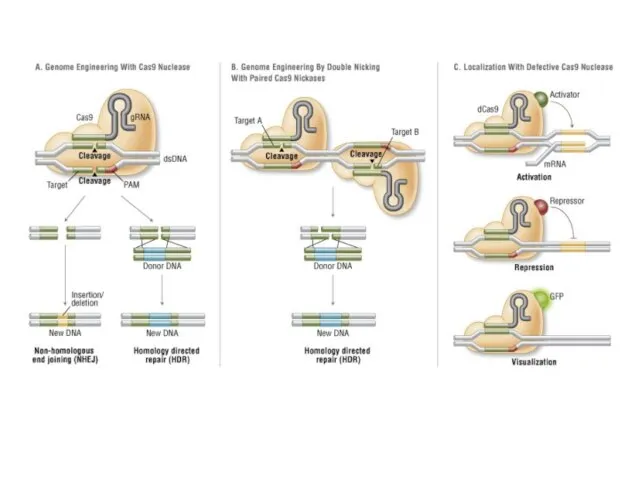
- 2. CRISPR Clustered Regularly Interspaced Short Palindromic Repeats Короткие палиндромные кластерные повторы, или CRISPR
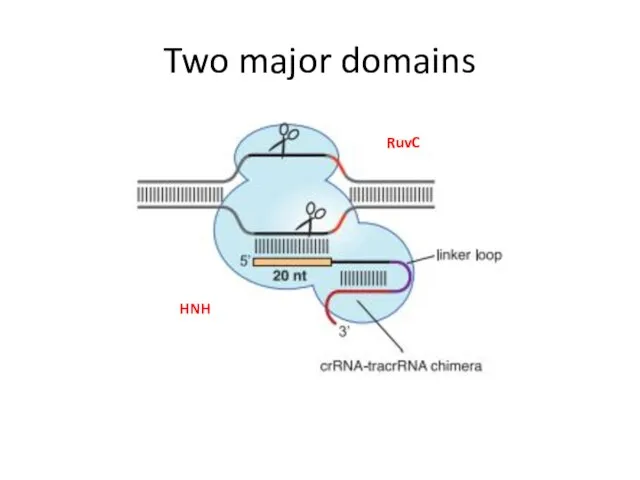
- 12. Two major domains RuvC HNH
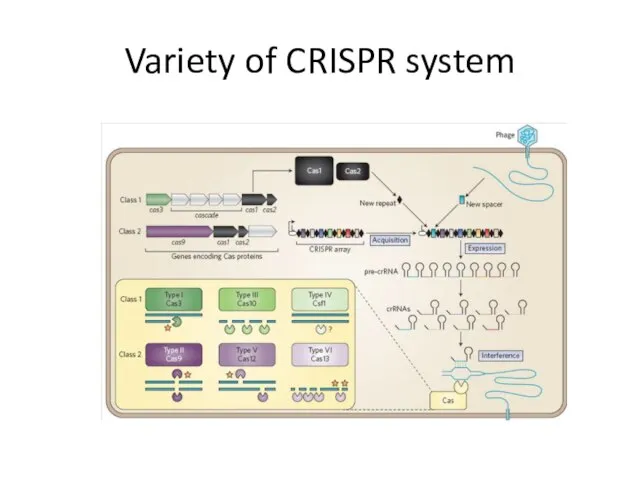
- 15. Variety of CRISPR system
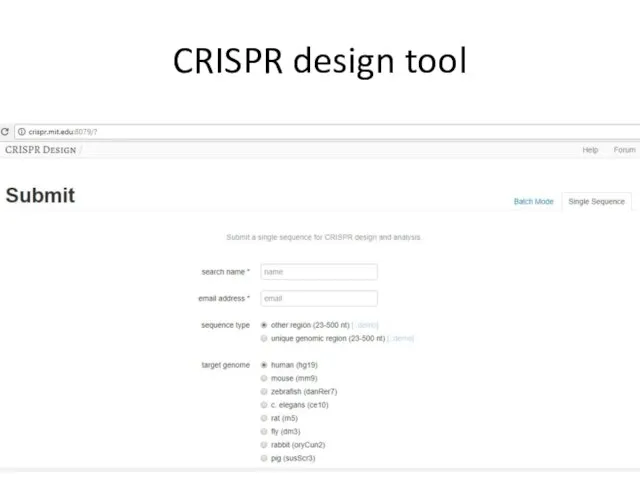
- 16. CRISPR design tool
- 17. W.-H. Wu et al., 2016. CRISPR repair reveals causative mutation in a preclinical model of retinitis
- 18. K. A. Schaefer et al., 2017. Unexpected mutations after CRISPR-Cas9 editing in vivo 117 insertions/delitions 1397
- 19. TALEN transcription activator-like nuclease
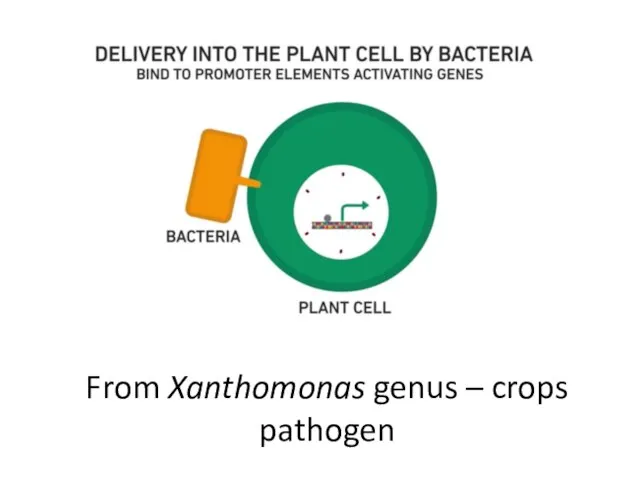
- 20. From Xanthomonas genus – crops pathogen
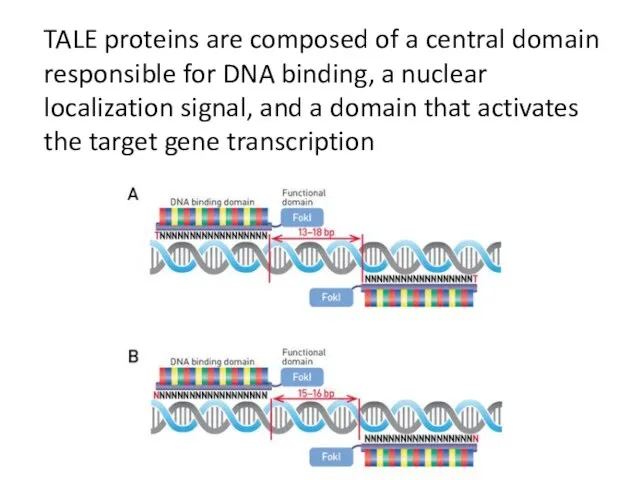
- 21. TALE proteins are composed of a central domain responsible for DNA binding, a nuclear localization signal,
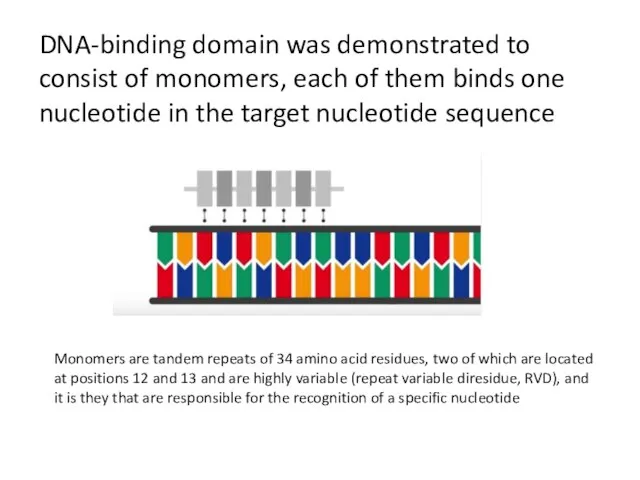
- 22. DNA-binding domain was demonstrated to consist of monomers, each of them binds one nucleotide in the
- 25. Скачать презентацию









 Здоровое сердце
Здоровое сердце Резективные методы лечения. (гингивэктомия, гемисекция, ампутация корня зуба)
Резективные методы лечения. (гингивэктомия, гемисекция, ампутация корня зуба) Железодифецитная анемия
Железодифецитная анемия Инфекция. Патогенные и вирулентные свойства бактерий
Инфекция. Патогенные и вирулентные свойства бактерий Bronchitis
Bronchitis Гипоталамический синдром пубертатного периода как органическое заболевание
Гипоталамический синдром пубертатного периода как органическое заболевание ВИЧ-инфекция.Туберкулез. Саркоидоз. Микозы
ВИЧ-инфекция.Туберкулез. Саркоидоз. Микозы Способности
Способности Деловое общение: специфика, формы, средства, функции
Деловое общение: специфика, формы, средства, функции Бүйрек қызметтері
Бүйрек қызметтері Правила проведения трансфузионной терапии
Правила проведения трансфузионной терапии Аускультация сердца
Аускультация сердца Організація та зміст роботи екстренної медичної допомоги. Зміст роботи станцій емд облік та аналіз їх діяльності
Організація та зміст роботи екстренної медичної допомоги. Зміст роботи станцій емд облік та аналіз їх діяльності Специальность сестринское дело. Промежуточная и итоговая аттестация
Специальность сестринское дело. Промежуточная и итоговая аттестация Trypanosoma Brucei
Trypanosoma Brucei Ацетонемический синдром
Ацетонемический синдром Здоровый образ жизни - путь к достижению высокого уровня здоровья
Здоровый образ жизни - путь к достижению высокого уровня здоровья Пульпит
Пульпит Дошкольная гигиена
Дошкольная гигиена Минералдар. Табиғи процестер
Минералдар. Табиғи процестер Патология органов речи
Патология органов речи Герпетическая инфекция
Герпетическая инфекция Осмотр пациента и оценка тяжести его состояния
Осмотр пациента и оценка тяжести его состояния Правила выписывания рецептов. Виды рецептурных бланков
Правила выписывания рецептов. Виды рецептурных бланков Общее недоразвитие речи у детей с нормальным слухом. Тест
Общее недоразвитие речи у детей с нормальным слухом. Тест Эпидемическое вирусное заболевание краснуха
Эпидемическое вирусное заболевание краснуха Жүрек, артериялар мен веналар құрылысы, жасқа сай ерекшеліктері
Жүрек, артериялар мен веналар құрылысы, жасқа сай ерекшеліктері Для чего же нужны эмоции?
Для чего же нужны эмоции?